5S3A
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1562205518 | | Descriptor: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.178 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z384468096 | | Descriptor: | N-[(4-phenyloxan-4-yl)methyl]acetamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0136 | | Descriptor: | (3S,4S)-4-(3-methoxyphenyl)oxane-3-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4E
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2301685688 | | Descriptor: | 1H-imidazole-5-carbonitrile, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5IDO
 
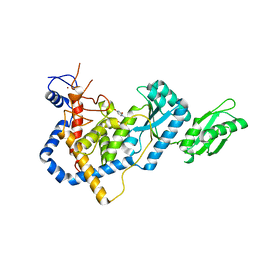 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5KAL
 
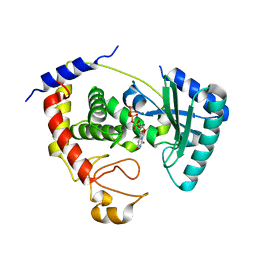 | | Terminal uridylyl transferase 4 from Trypanosoma brucei with bound UTP and UpU | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*U)-3'), RNA uridylyltransferase 4, ... | | Authors: | Stagno, J.R, Luecke, H, Afasizhev, R. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
2IKF
 
 | |
4J68
 
 | |
8BQU
 
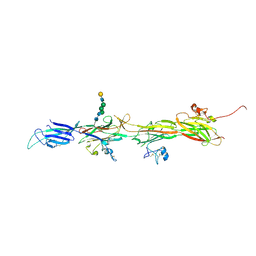 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
6R37
 
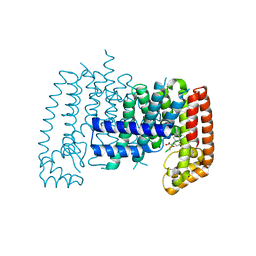 | | T. brucei FPPS in complex with 2-(5-chlorobenzo[b]thiophen-3-yl)acetic acid | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)acetic acid, DIMETHYL SULFOXIDE, Farnesyl pyrophosphate synthase | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Trypanosoma brucei FPPS by Fragment-based drug discovery
Thesis, 2019
|
|
5SA8
 
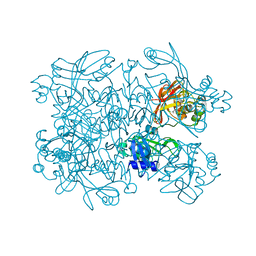 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z68299550 | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
2K2Y
 
 | |
6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
6T68
 
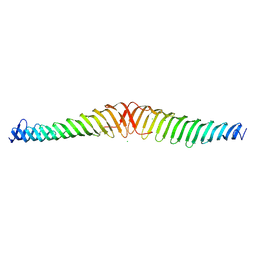 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
3OTX
 
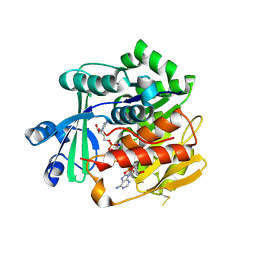 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Inhibitor AP5A | | Descriptor: | Adenosine kinase, putative, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of T. b. rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation
Plos Negl Trop Dis, 5, 2011
|
|
6T4R
 
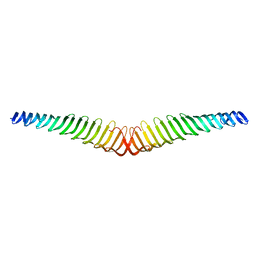 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
2K2Z
 
 | |
6NL1
 
 | | Structure of T. brucei MERS1 protein in its apo form | | Descriptor: | Mitochondrial edited mRNA stability factor 1, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
5KRE
 
 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
6R04
 
 | | T. cruzi FPPS | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
6R08
 
 | | T. cruzi FPPS in complex with 3-(carboxymethyl)-5,7-dichloro-1H-indole-2-carboxylic acid | | Descriptor: | 3-(carboxymethyl)-5,7-dichloro-1H-indole-2-carboxylic acid, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
6R06
 
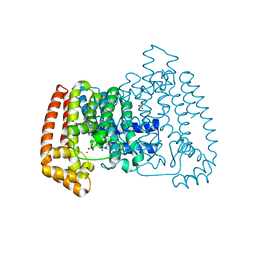 | | T. CRUZI FPPS IN COMPLEX WITH (3S,4S)-4-(3,4-dimethylphenoxy)-1-(prop-2-yn-1-yl)piperidin-3-ol | | Descriptor: | (3~{S},4~{S})-4-(3,4-dimethylphenoxy)-1-prop-2-ynyl-piperidin-3-ol, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Wolfgang, J. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
6R0B
 
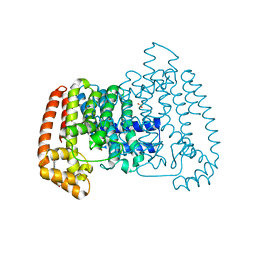 | | T. cruzi FPPS in complex with 3-((4-(5-chlorobenzo[d]thiazol-2-yl)piperazin-1-yl)methyl)-1H-indol-5-ol | | Descriptor: | 3-[[4-(5-chloranyl-1,3-benzothiazol-2-yl)piperazin-1-yl]methyl]-1~{H}-indol-5-ol, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
6R05
 
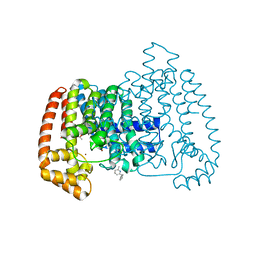 | | T. CRUZI FPPS IN COMPLEX WITH N-BENZYL-6-METHYLPYRIDIN-2-AMINE | | Descriptor: | 6-methyl-~{N}-(phenylmethyl)pyridin-2-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
