8VML
 
 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VCF
 
 | |
7MAW
 
 | |
7MAV
 
 | |
5I5W
 
 | |
8VOB
 
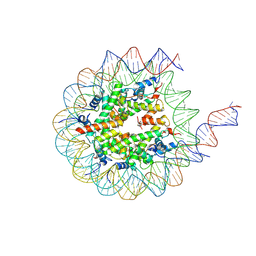 | | H3K36me3-modified nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5VIG
 
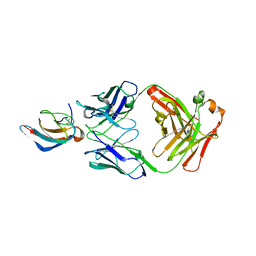 | | Crystal structure of anti-Zika antibody Z006 bound to Zika virus envelope protein DIII | | Descriptor: | CITRATE ANION, Fab heavy chain, Fab light chain, ... | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
7MAX
 
 | |
8VS0
 
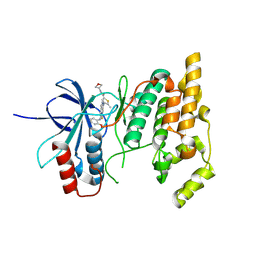 | |
5IP1
 
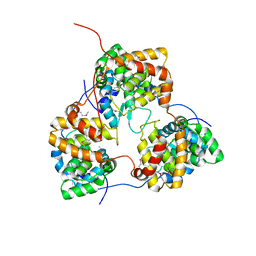 | | Tomato spotted wilt tospovirus nucleocapsid protein | | Descriptor: | Nucleoprotein | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
8VW9
 
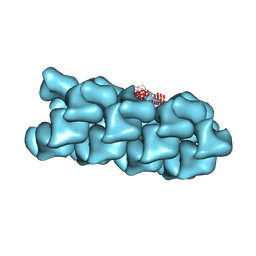 | | CryoEM Structure of a FtsH Helical Assembly in the Presence of ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, ... | | Authors: | Li, Y, Zhu, J, Zhang, Z, Wang, F, Egelman, E.H, Tezcan, F.A. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Transforming an ATP-dependent enzyme into a dissipative, self-assembling system.
Nat.Chem.Biol., 21, 2025
|
|
6W8T
 
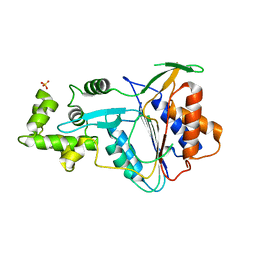 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
8VWC
 
 | | CryoEM Structure of a FtsH Helical Assembly in the Aged State | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, ... | | Authors: | Li, Y, Zhu, J, Zhang, Z, Wang, F, Egelman, E.H, Tezcan, F.A. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Transforming an ATP-dependent enzyme into a dissipative, self-assembling system.
Nat.Chem.Biol., 21, 2025
|
|
5VKB
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and argininamide | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININEAMIDE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
7MB0
 
 | |
6W0H
 
 | | Closed-gate KcsA soaked in 5mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
5VP8
 
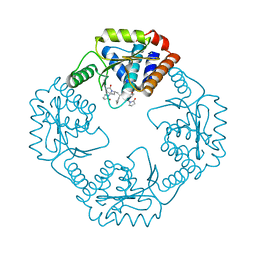 | | I38T mutant of 2009 H1N1 PA Endonuclease | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
8VVY
 
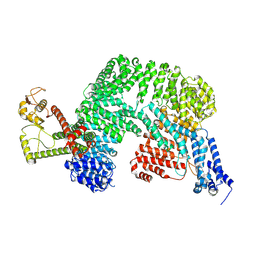 | | Human Cullin-1 in complex with CAND2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 2 | | Authors: | Kenny, S, Liu, X, Das, C. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanisms of CAND2 in regulating SCF ubiquitin ligases.
Nat Commun, 16, 2025
|
|
7M6N
 
 | |
5VL4
 
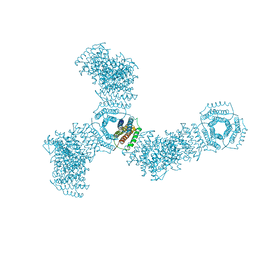 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
7M6Q
 
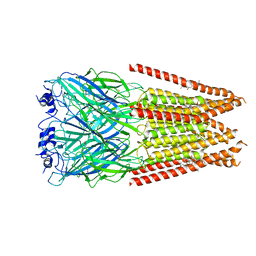 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
8VWY
 
 | | Complex structure of mouse C1ql3 with BAI3 | | Descriptor: | Adhesion G protein-coupled receptor B3, CALCIUM ION, Complement C1q-like protein 3 | | Authors: | Miao, Y, Sudhof, T.C. | | Deposit date: | 2024-02-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structure of the complex of C1q-like 3 protein with adhesion-GPCR BAI3.
Commun Biol, 8, 2025
|
|
7M6R
 
 | |
6WA8
 
 | |
5I7M
 
 | |
