2L9E
 
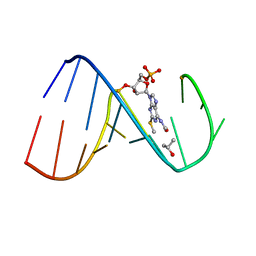 | | Solution Structure of the human Anti-codon Stem and loop(hASL) of transfer RNA Lysine 3 (tRNALys3) | | Descriptor: | RNA (5'-R(*GP*CP*AP*GP*AP*CP*UP*(70U)P*UP*UP*(12A)P*AP*UP*CP*UP*GP*C)-3') | | Authors: | Vendeix, F.A.P, Murphy IV, F.V, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the human Anti-codon Stem and loop(hASL) of transfer RNA Lysine 3 (tRNALys3)
To be Published
|
|
5IRR
 
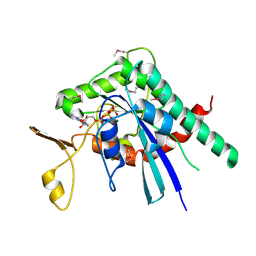 | | Crystal structure of Septin GTPase domain from Chlamydomonas reinhardtii | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Septin-like protein | | Authors: | Pinto, A.P.A, Pereira, H.M, Navarro, M.V.A.S, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Filaments and fingers: Novel structural aspects of the single septin from Chlamydomonas reinhardtii.
J. Biol. Chem., 292, 2017
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T1H
 
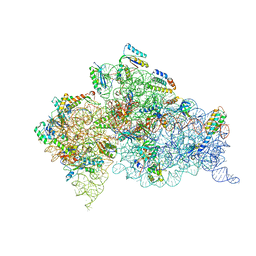 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
4WCT
 
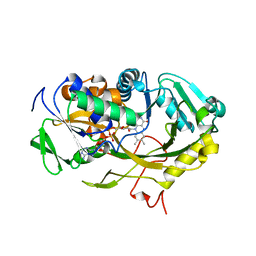 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
6D6Y
 
 | |
4FA3
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid (86) | | Descriptor: | (3R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
8PAZ
 
 | | OXIDIZED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-24 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
6FAD
 
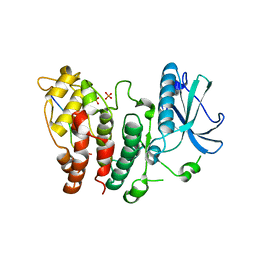 | | SR protein kinase 1 (SRPK1) in complex with the RGG-box of HSV1 ICP27 | | Descriptor: | PHOSPHATE ION, SRSF protein kinase 1, mRNA export factor | | Authors: | Tunnicliffe, R.B, Levy, C, Mould, A.P, Mckenzie, E.A, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.
Mbio, 10, 2019
|
|
1G0X
 
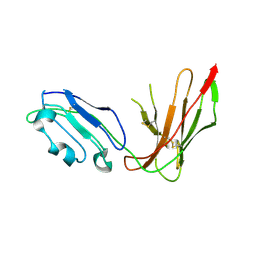 | |
4OY9
 
 | | Crystal structure of human P-Cadherin EC1-EC2 in closed conformation | | Descriptor: | CALCIUM ION, Cadherin-3 | | Authors: | Dalle Vedove, A, Lucarelli, A.P, Nardone, V, Matino, A, Parisini, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The X-ray structure of human P-cadherin EC1-EC2 in a closed conformation provides insight into the type I cadherin dimerization pathway.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1VM1
 
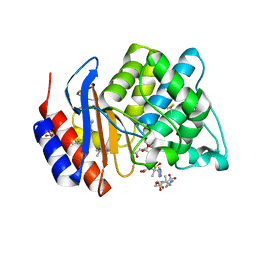 | | STRUCTURE OF SHV-1 BETA-LACTAMASE INHIBITED BY TAZOBACTAM | | Descriptor: | ACRYLIC ACID, BETA-LACTAMASE SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | Kuzin, A.P, Nukaga, M, Nukaga, Y, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibition of the SHV-1 beta-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam.
Biochemistry, 40, 2001
|
|
4OPH
 
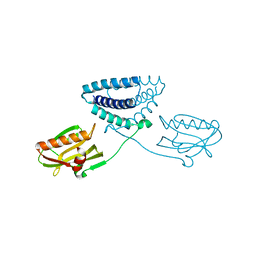 | | X-ray structure of full-length H6N6 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
1W2Z
 
 | | PSAO and Xenon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMINE OXIDASE, COPPER CONTAINING, ... | | Authors: | Duff, A.P, Trambaiolo, D.M, Cohen, A.E, Ellis, P.J, Juda, G.A, Shepard, E.M, Langley, D.B, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-Binding Sites in Copper Amine Oxidases.
J.Mol.Biol., 344, 2004
|
|
3KVP
 
 | | Crystal Structure of Uncharacterized protein ymzC Precursor from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR378A | | Descriptor: | ACETIC ACID, Uncharacterized protein ymzC | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Afonine, P, Fang, F, Xiao, R, Cunningham, K, Ma, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR378A
To be Published
|
|
7NEE
 
 | | Inhibitor Complex with Thrombin Activatable Fibrinolysis inhibitor (TAFIa) | | Descriptor: | (1R,3S)-3-(4-ammoniobutyl)-1-benzyl-1,4-azaphosphinan-1-ium-3-carboxylate 4,4-dioxide, Carboxypeptidase B2, ZINC ION | | Authors: | Brown, D.G, Schaffner, A.P, Gloanec, P, Raimbaud, E, Vuillard, L.M. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphinanes and Azaphosphinanes as Potent and Selective Inhibitors of Activated Thrombin-Activatable Fibrinolysis Inhibitor (TAFIa).
J.Med.Chem., 64, 2021
|
|
7NEU
 
 | | Inhibitor Complex with Thrombin Activatable Fibrinolysis Inhibitor (TAFIa) | | Descriptor: | (1R,3S)-3-(4-ammoniobutyl)-1-(4-fluoro-2-(1-methyl-1H-imidazol-5-yl)benzyl)-1,4-azaphosphinan-1-ium-3-carboxylate 4,4-dioxide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brown, D.G, Schaffner, A.P, Vuillard, L.M, Gloanec, P, Raimbauld, E. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphinanes and Azaphosphinanes as Potent and Selective Inhibitors of Activated Thrombin-Activatable Fibrinolysis Inhibitor (TAFIa).
J.Med.Chem., 64, 2021
|
|
3L6I
 
 | | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542 | | Descriptor: | SODIUM ION, Uncharacterized lipoprotein yceB | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Chen, C.X, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542
To be Published
|
|
8G4K
 
 | |
8OH4
 
 | |
1URK
 
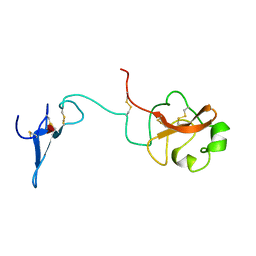 | | SOLUTION STRUCTURE OF THE AMINO TERMINAL FRAGMENT OF UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | PLASMINOGEN ACTIVATOR, alpha-L-fucopyranose | | Authors: | Hansen, A.P, Petros, A.M, Meadows, R.P, Nettesheim, D.G, Mazar, A.P, Olejniczak, E.T, Xu, R.X, Pederson, T.M, Henkin, J, Fesik, S.W. | | Deposit date: | 1994-01-10 | | Release date: | 1995-05-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator.
Biochemistry, 33, 1994
|
|
7R7G
 
 | |
7R7F
 
 | |
7R7E
 
 | |
