4CI8
 
 | |
5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
4E5Z
 
 | |
4E54
 
 | |
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XXQ
 
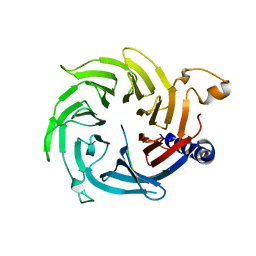 | |
5Y1U
 
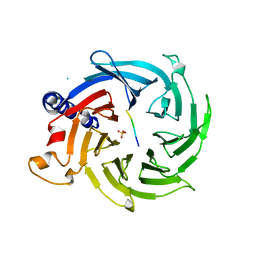 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
3GFC
 
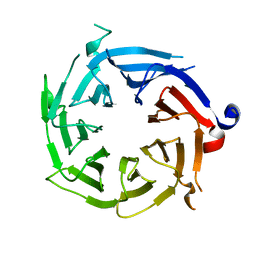 | | Crystal Structure of Histone-binding protein RBBP4 | | Descriptor: | Histone-binding protein RBBP4 | | Authors: | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
3IKO
 
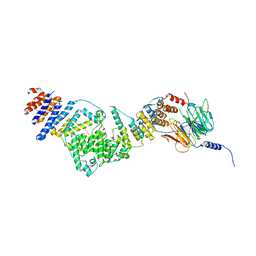 | | Crystal structure of the heterotrimeric Sec13-Nup145C-Nup84 nucleoporin complex | | Descriptor: | Nucleoporin NUP145C, Nucleoporin NUP84, Protein transport protein SEC13 | | Authors: | Nagy, V, Hsia, K.-C, Debler, E.W, Davenport, A, Blobel, G, Hoelz, A. | | Deposit date: | 2009-08-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a trimeric nucleoporin complex reveals alternate oligomerization states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
6B20
 
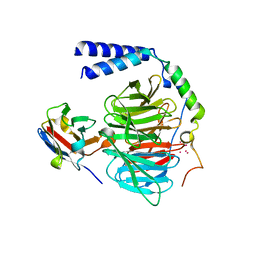 | | Crystal structure of a complex between G protein beta gamma dimer and an inhibitory Nanobody regulator | | Descriptor: | CHLORIDE ION, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gulati, S, Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor.
Nat Commun, 9, 2018
|
|
3JRO
 
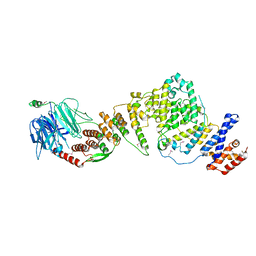 | | NUP84-NUP145C-SEC13 edge element of the NPC lattice | | Descriptor: | Fusion Protein of Protein Transport Protein SEC13 and Nucleoporin NUP145, Nucleoporin NUP84 | | Authors: | Brohawn, S.G, Schwartz, T.U. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-27 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6BCX
 
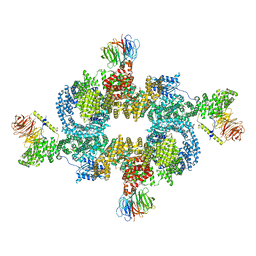 | | mTORC1 structure refined to 3.0 angstroms | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Nature, 552, 2017
|
|
6BX3
 
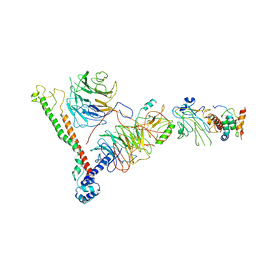 | | Structure of histone H3k4 methyltransferase | | Descriptor: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | Authors: | Skiniotis, G, Qu, Q.H. | | Deposit date: | 2017-12-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
6BM0
 
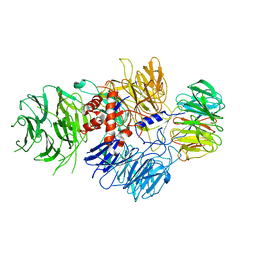 | | Cryo-EM structure of human CPSF-160-WDR33 complex at 3.8 A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2017-11-12 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BLY
 
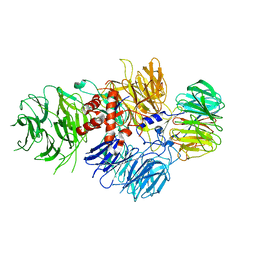 | | Cryo-EM structure of human CPSF-160-WDR33 complex at 3.36A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2017-11-12 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C23
 
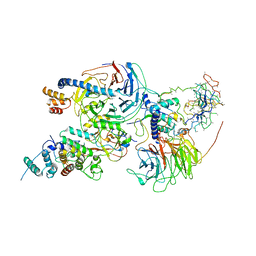 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
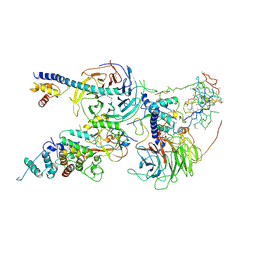 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6BW3
 
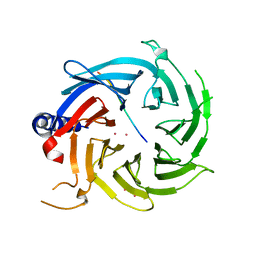 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6BW4
 
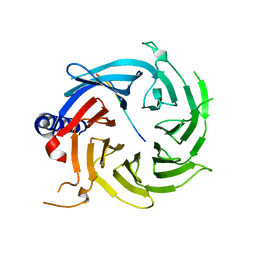 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6CRK
 
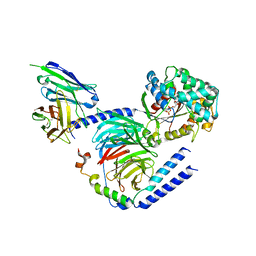 | | Heterotrimeric G-protein in complex with an antibody fragment | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Maeda, S, Dawson, R, Kobilka, B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an antibody fragment that stabilizes GPCR/G-protein complexes.
Nat Commun, 9, 2018
|
|
6CKC
 
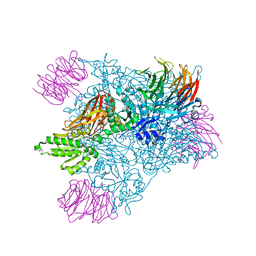 | | Structure of PRMT5:MEP50 in complex with LLY-283, a potent and selective inhibitor of PRMT5, with antitumor activity | | Descriptor: | 7-[(5R)-5-C-phenyl-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Antonysamy, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity.
ACS Med Chem Lett, 9, 2018
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
4FHM
 
 | |
