1KCL
 
 | | Bacillus ciruclans strain 251 Cyclodextrin glycosyl transferase mutant G179L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cyclodextrin glycosyltransferase, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1TX8
 
 | | Bovine Trypsin complexed with AMSO | | Descriptor: | 4-(METHYLSULFONYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, Trypsinogen | | Authors: | Mesecar, A.D. | | Deposit date: | 2004-07-02 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis, and evaluation of oxyanion-hole selective inhibitor substituents for the S1 subsite of factor Xa
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
2OUA
 
 | | Crystal Structure of Nocardiopsis Protease (NAPase) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic exploration of Acid resistance: kinetic stability facilitates evolution of extremophilic behavior
J.Mol.Biol., 368, 2007
|
|
1UHL
 
 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
2MZV
 
 | |
2P8H
 
 | | Crystal structure of human beta secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-{(1S,2S)-1-BENZYL-2-HYDROXY-2-[(4S)-1,2,2-TRIMETHYL-5-OXOIMIDAZOLIDIN-4-YL]ETHYL}-N'-[(1R)-1-(4-FLUOROPHENYL)ETHYL]-5-[METHYL(METHYLSULFONYL)AMINO]ISOPHTHALAMIDE | | Authors: | Munshi, S. | | Deposit date: | 2007-03-22 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of 2,3,5-Substituted Imidazolidin-4-one Inhibitors of BACE-1.
Chemmedchem, 2, 2007
|
|
2LTB
 
 | | Wild-type FAS1-4 | | Descriptor: | Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Underhaug, J, Nielsen, N, Runager, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mutation in transforming growth factor beta induced protein associated with granular corneal dystrophy type 1 reduces the proteolytic susceptibility through local structural stabilization.
Biochim.Biophys.Acta, 1834, 2013
|
|
1UPV
 
 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
2PFE
 
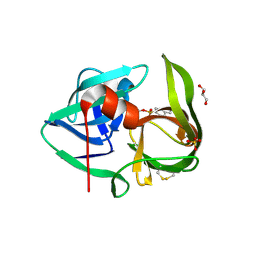 | | Crystal Structure of Thermobifida fusca Protease A (TFPA) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Alkaline serine protease, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Mesophile versus Thermophile: Insights Into the Structural Mechanisms of Kinetic Stability
J.Mol.Biol., 370, 2007
|
|
2PM5
 
 | |
2PKS
 
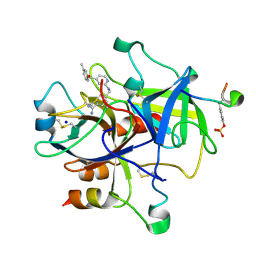 | | Thrombin in complex with inhibitor | | Descriptor: | 4-({[4-(3-METHYLBENZOYL)PYRIDIN-2-YL]AMINO}METHYL)BENZENECARBOXIMIDAMIDE, Hirudin, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2007-04-18 | | Release date: | 2008-04-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and biological evaluation of thrombin inhibitors based on a pyridine scaffold.
Org.Biomol.Chem., 5, 2007
|
|
2PM1
 
 | |
1UPW
 
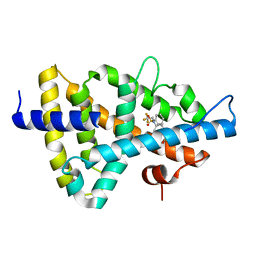 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
2M0K
 
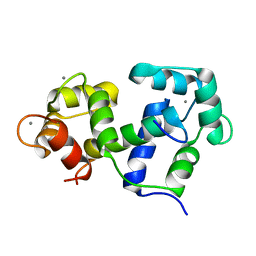 | |
2PM4
 
 | | Human alpha-defensin 1 (multiple Arg->Lys mutant) | | Descriptor: | Neutrophil defensin 1 (HNP-1) (HP-1) (HP1) (Defensin, alpha 1) | | Authors: | Lubkowski, J, Pazgier, M, Lu, W. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs.
J.Biol.Chem., 282, 2007
|
|
2MMW
 
 | |
2M0J
 
 | |
2PLZ
 
 | | Arg-modified human beta-defensin 1 (HBD1) | | Descriptor: | Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M, Lu, W. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs.
J.Biol.Chem., 282, 2007
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
7EL7
 
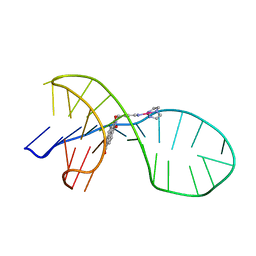 | | NMR solution structure of the 1:1 complex of a quadruplex-duplex hybrid MYT1L and a platinum(II) ligand L1Pt(dien) | | Descriptor: | G-quadruplex DNA MYT1L, Pt(diethylenetriamine)(2-(pyridin-4-ylmethyl)benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Liu, W, Mao, Z.-W. | | Deposit date: | 2021-04-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spatial Matching Selectivity and Solution Structure of Organic-Metal Hybrid to Quadruplex-Duplex Hybrid.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5YE1
 
 | |
5Y66
 
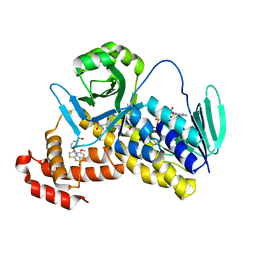 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
8F7V
 
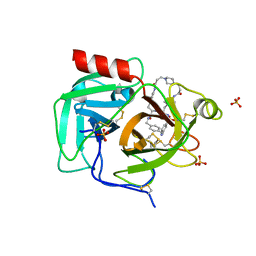 | | Macrocyclic plasmin inhibitor | | Descriptor: | (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(benzenesulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, GLYCEROL, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-11-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and structural characterization of new macrocyclicplasmin inhibitors
Chemmedchem, 2023
|
|
5YGP
 
 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
