7BED
 
 | |
7BCY
 
 | |
2KTU
 
 | |
2KDP
 
 | | Solution Structure of the SAP30 zinc finger motif | | Descriptor: | Histone deacetylase complex subunit SAP30, ZINC ION | | Authors: | He, Y, Imhoff, R, Sahu, A, Radhakrishnan, I. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition
Nucleic Acids Res., 37, 2009
|
|
4PJW
 
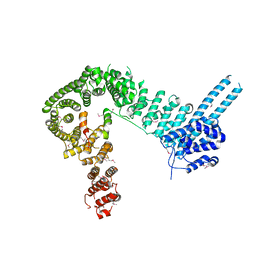 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PL7
 
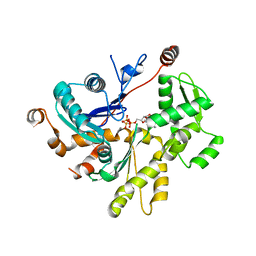 | |
4PJT
 
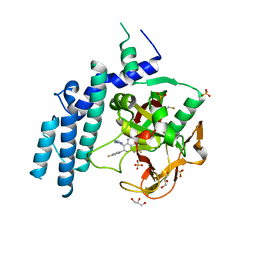 | | Structure of PARP1 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Arakaki, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4PJU
 
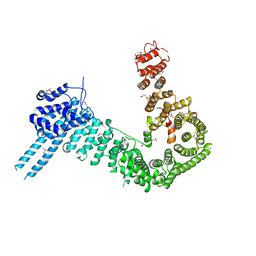 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PL8
 
 | |
4PK7
 
 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7EPZ
 
 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
4RXQ
 
 | | The structure of GTP-dUTP-bound SAMHD1 | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RXO
 
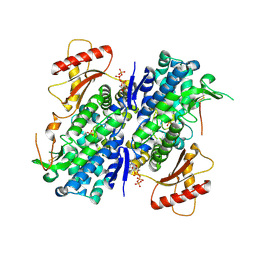 | | The structure of GTP-bound SAMHD1 | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7EQ7
 
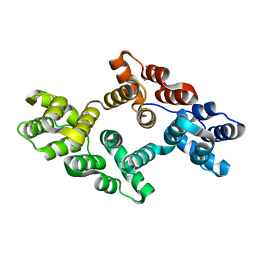 | |
4RV6
 
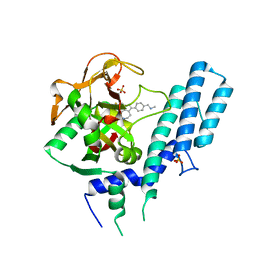 | |
2KIZ
 
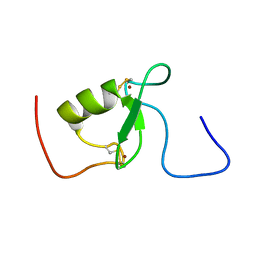 | | Solution structure of Arkadia RING-H2 finger domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Kandias, N.G, Chasapis, C.T, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2013-03-20 | | Method: | SOLUTION NMR | | Cite: | NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.
Proteins, 80, 2012
|
|
4TO0
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP-dCTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TO3
 
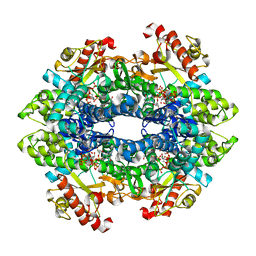 | | Structural basis of cellular dNTP regulation, SAMHD1-dGTP-dGTP-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KMD
 
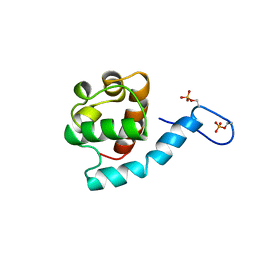 | | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to co-activator CBP | | Descriptor: | Protein C-ets-1 | | Authors: | Nelson, M.L, Kang, H, Lee, G.M, Blaszczak, A.G, Lau, D.K.W, McIntosh, L.P, Graves, B.J. | | Deposit date: | 2009-07-27 | | Release date: | 2010-05-05 | | Last modified: | 2012-03-21 | | Method: | SOLUTION NMR | | Cite: | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KTV
 
 | |
7C2Z
 
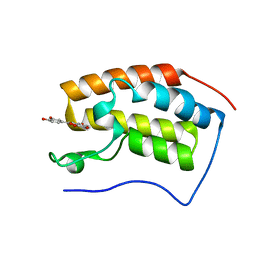 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
2KTB
 
 | |
2KNV
 
 | |
4O78
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with GW612286X | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
2KRM
 
 | |
