7SXR
 
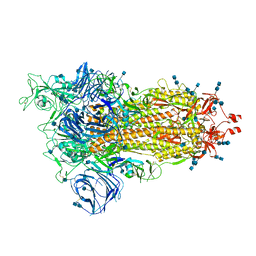 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXV
 
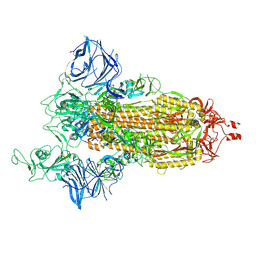 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417N mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXU
 
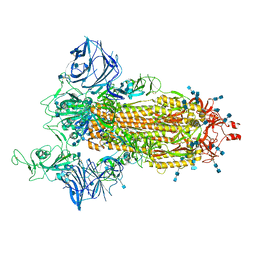 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXX
 
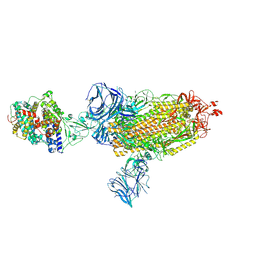 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
6QD8
 
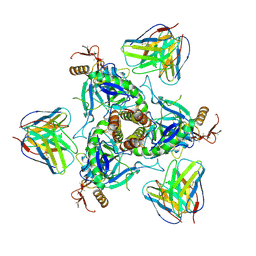 | | EM structure of a EBOV-GP bound to 4M0368 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | rVSV-ZEBOV induces a polyclonal and convergent B cell response with potent Ebola virus-neutralizing antibodies
Nat.Med. (N.Y.), 2019
|
|
2M8O
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in DPC | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
8BCZ
 
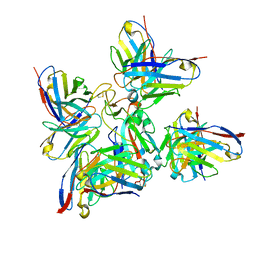 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
6LTG
 
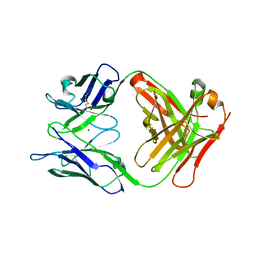 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
2M8M
 
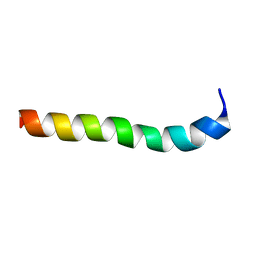 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
7WPH
 
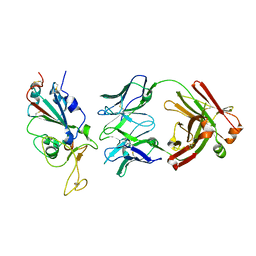 | | SARS-CoV2 RBD bound to Fab06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
5C0S
 
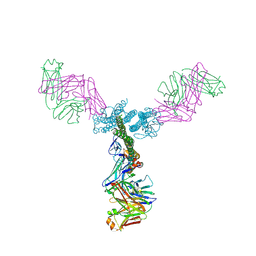 | | Crystal structure of a generation 4 influenza hemagglutinin stabilized stem in complex with the broadly neutralizing antibody CR6261 | | Descriptor: | CR6261 antibody heavy chain, CR6261 antibody light chain, Hemagglutinin, ... | | Authors: | Boyington, J.C, Kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
5NHR
 
 | |
2MTW
 
 | | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit into HLA-DR 1*03 molecules induces antibody production and protection | | Descriptor: | Erythrocyte-binding antigen 175 | | Authors: | Cifuentes, G, Salazar, L, Vargas, L, Parra, C, Vanegas, M, Cortes, J, Sandoval, M, Patarroyo, M.E. | | Deposit date: | 2014-09-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit HLA-DR 1*03 molecules induces antibody production and protection
To be Published
|
|
5NGV
 
 | |
4G6K
 
 | |
1EZV
 
 | | STRUCTURE OF THE YEAST CYTOCHROME BC1 COMPLEX CO-CRYSTALLIZED WITH AN ANTIBODY FV-FRAGMENT | | Descriptor: | 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CYTOCHROME B, CYTOCHROME C1, ... | | Authors: | Hunte, C, Koepke, J, Lange, C, Rossmanith, T, Michel, H. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment.
Structure Fold.Des., 8, 2000
|
|
1HH6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
4DN4
 
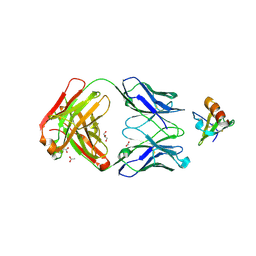 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
7X2H
 
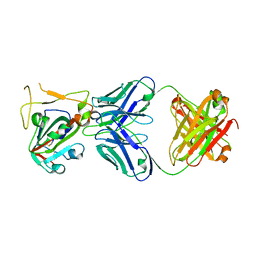 | |
2QSC
 
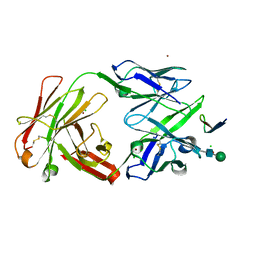 | | Crystal structure analysis of anti-HIV-1 V3-Fab F425-B4e8 in complex with a V3-peptide | | Descriptor: | CHLORIDE ION, Envelope glycoprotein gp120, Fab F425-B4e8, ... | | Authors: | Bell, C.H, Schiefner, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization.
J.Mol.Biol., 375, 2008
|
|
6HX4
 
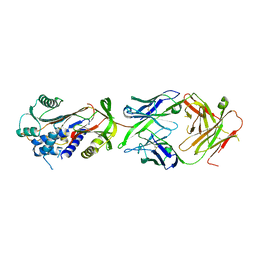 | | Fab fragment of a native monomer-selective antibody in complex with alpha-1-antitrypsin | | Descriptor: | Alpha-1-antitrypsin, Fab 1D9 heavy chain, Fab 1D9 light chain | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
6EMJ
 
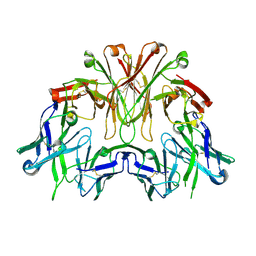 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | SODIUM ION, fAB heavy chain, fAb light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELL
 
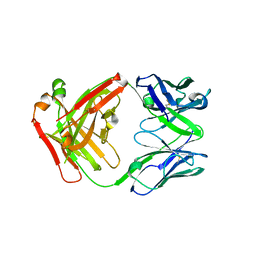 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
7Y0V
 
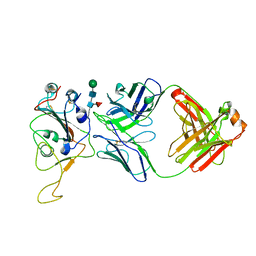 | | The co-crystal structure of BA.1-RBD with Fab-5549 | | Descriptor: | 5549-Fab, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Q4Q
 
 | | Magacizumab Fab fragment in complex with human LRG1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRG1 epitope, Magacizumab heavy chain, ... | | Authors: | Gutierrez-Fernandez, J, Luecke, H. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of human LRG1 recognition by Magacizumab, a humanized monoclonal antibody with therapeutic potential.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
