4V2R
 
 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | Descriptor: | PHENYLALANINE AMINOMUTASE (L-BETA-PHENYLALANINE FORMING) | | Authors: | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ironing out their differences: dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase.
ACS Chem. Biol., 10, 2015
|
|
7LS0
 
 | | Structure of the Human ALK GRD bound to AUG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor fused with ALK and LTK ligand 2, CITRIC ACID | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
6CBP
 
 | |
7LI9
 
 | |
5VC1
 
 | | Crystal structure of L-selectin lectin/EGF domains | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wedepohl, S, Dernedde, J, Vahedi-Faridi, A, Tauber, R, Saenger, W, Bulut, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Reducing Macro- and Microheterogeneity of N-Glycans Enables the Crystal Structure of the Lectin and EGF-Like Domains of Human L-Selectin To Be Solved at 1.9 angstrom Resolution.
Chembiochem, 18, 2017
|
|
5VCS
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with Bound Acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7LI7
 
 | |
4WCU
 
 | | PDE4 complexed with inhibitor | | Descriptor: | MAGNESIUM ION, N-benzyl-2-{6-[(3,5-dichloropyridin-4-yl)acetyl]-2,3-dimethoxyphenoxy}acetamide, ZINC ION, ... | | Authors: | Sorensen, M.D. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Early Clinical Development of 2-{6-[2-(3,5-Dichloro-4-pyridyl)acetyl]-2,3-dimethoxyphenoxy}-N-propylacetamide (LEO 29102), a Soft-Drug Inhibitor of Phosphodiesterase 4 for Topical Treatment of Atopic Dermatitis.
J. Med. Chem., 57, 2014
|
|
7LIA
 
 | |
7LI6
 
 | | apo SERT reconstituted in lipid nanodisc in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Yang, D, Gouaux, E. | | Deposit date: | 2021-01-26 | | Release date: | 2021-12-15 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Illumination of serotonin transporter mechanism and role of the allosteric site.
Sci Adv, 7, 2021
|
|
7M74
 
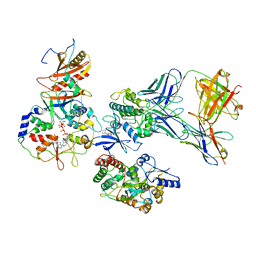 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
4UZU
 
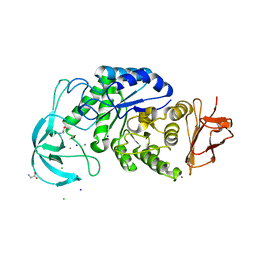 | | Three-dimensional structure of a variant `Termamyl-like' Geobacillus stearothermophilus alpha-amylase at 1.9 A resolution | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Anderson, C, Borchert, T.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of a Variant `Termamyl-Like' Geobacillus Stearothermophilus Alpha-Amylase at 1.9 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7LI8
 
 | |
4UQQ
 
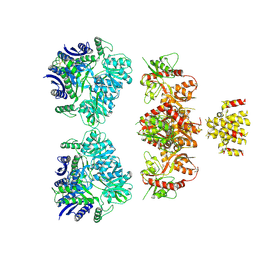 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UY8
 
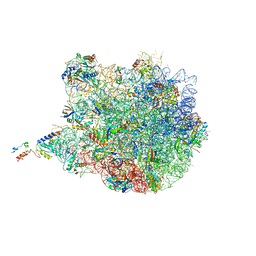 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Bischoff, L, Berninghausen, O, Beckmann, R. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
5IRE
 
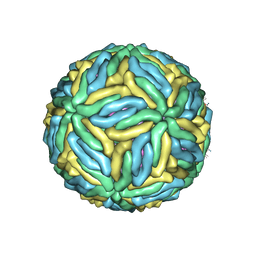 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
4W7L
 
 | |
5IIB
 
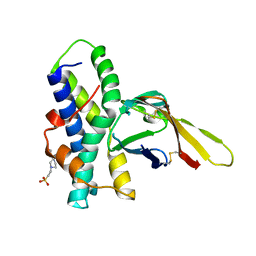 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | Authors: | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
4UUP
 
 | | Reconstructed ancestral trichomonad malate dehydrogenase in complex with NADH, SO4, and PO4 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
4UW4
 
 | | Human galectin-7 in complex with a galactose based dendron D2-1. | | Descriptor: | DENDRON D2-1, GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
4UY5
 
 | |
5IKR
 
 | | The Structure of Mefenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
5VIY
 
 | |
5VN4
 
 | |
