7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
3MUF
 
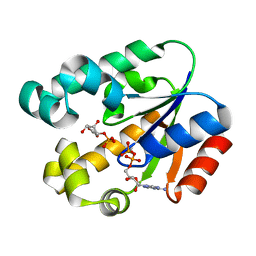 | | Shikimate kinase from Helicobacter pylori in complex with shikimate-3-phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE-3-PHOSPHATE, Shikimate kinase | | Authors: | Cheng, W.C, Chen, T.J, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-05-03 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Helicobacter pylori shikimate kinase reveal a selective inhibitor-induced-fit mechanism
Plos One, 7, 2012
|
|
6FF6
 
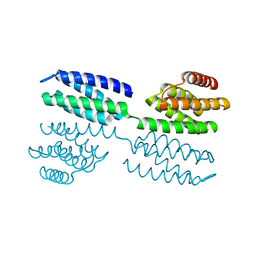 | | Crystal structure of novel repeat protein BRIC1 | | Descriptor: | BRIC1 | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A.N. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
5VN3
 
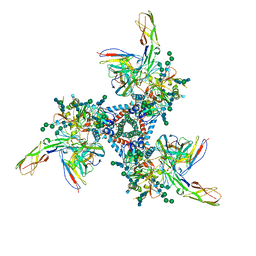 | | Cryo-EM model of B41 SOSIP.664 in complex with soluble CD4 (D1-D2) and fragment antigen binding variable domain of 17b | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
3MYR
 
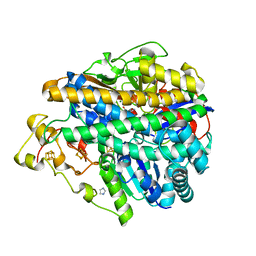 | | Crystal structure of [NiFe] hydrogenase from Allochromatium vinosum in its Ni-A state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Hydrogenase (NiFe) small subunit HydA, ... | | Authors: | Ogata, H, Kellers, P, Lubitz, W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A state).
J.Mol.Biol., 402, 2010
|
|
6G1X
 
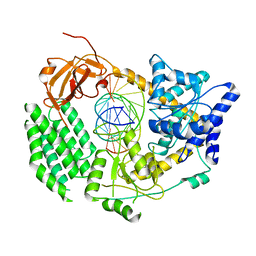 | | CryoEM structure of the MDA5-dsRNA filament with 91-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
5SW3
 
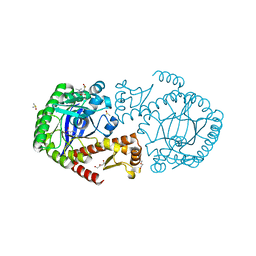 | | Crystal Structure of TGT in complex with 3-Pyridinecarboxylic acid, 6-(dimethylamino) | | Descriptor: | 6-(dimethylamino)pyridine-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2016-08-08 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
7BDV
 
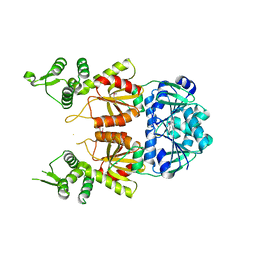 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
5OOK
 
 | | Structure of A. marina Phycocyanin contains overlapping isoforms | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phycocyanin, ... | | Authors: | Bar-Zvi, S, Lahav, A, Blankenship, E.R, Adir, N. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural heterogeneity leads to functional homogeneity in A. marina phycocyanin.
Biochim. Biophys. Acta, 1859, 2018
|
|
6FML
 
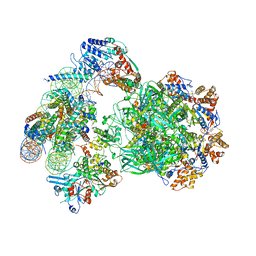 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
7BBW
 
 | |
7BBX
 
 | | Neisseria gonorrhoeae transaldolase, variant K8A | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.M, Tittmann, K. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
7BEV
 
 | |
7F7N
 
 | |
3N3Y
 
 | | Crystal structure of Thymidylate Synthase X (ThyX) from Helicobacter pylori with FAD and dUMP at 2.31A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Wang, K, Wang, Q, Chen, J, Chen, L, Jiang, H, Shen, X. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structure, Enzymatic Characterization and Inhibitor Discovery of Thymidylate Synthase X (ThyX) from Helicobacter pylori Strain SS1
To be Published
|
|
3JSF
 
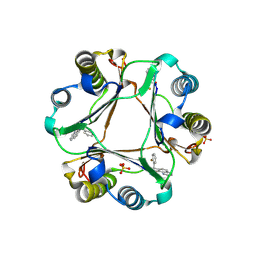 | |
5VN8
 
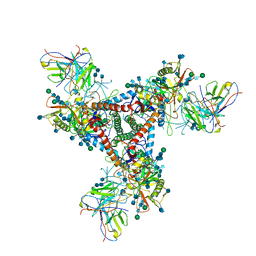 | | Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B, Cottrell, C.A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
7BO9
 
 | |
7BO8
 
 | | A hexameric de novo coiled-coil assembly: CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F. | | Descriptor: | 1,2-ETHANEDIOL, CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F, OXAMIC ACID | | Authors: | Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Coiled-Coil Assemblies Accommodate Multiple Aromatic Residues.
Biomacromolecules, 22, 2021
|
|
7BOA
 
 | |
6K3L
 
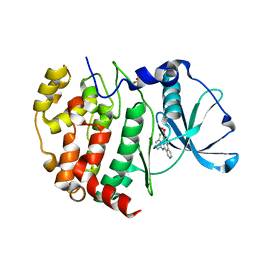 | | Crystal structure of CX-4945 bound Cka1 from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CMGC/CK2 protein kinase, SULFATE ION | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
3NKA
 
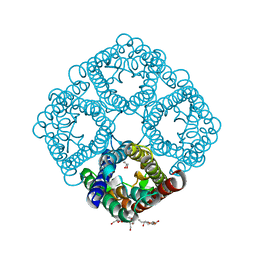 | | Crystal structure of AqpZ H174G,T183F | | Descriptor: | Aquaporin Z, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JAV
 
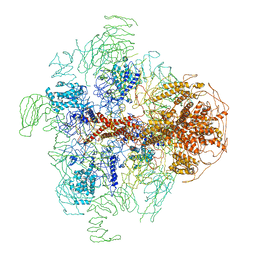 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
6G1S
 
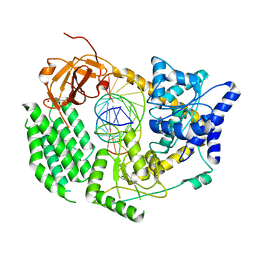 | | CryoEM structure of the MDA5-dsRNA filament with 87-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
