2HTE
 
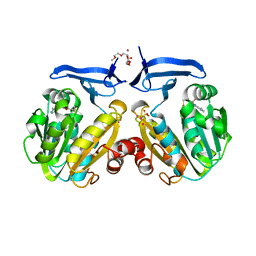 | | The crystal structure of spermidine synthase from p. falciparum in complex with 5'-methylthioadenosine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2FDS
 
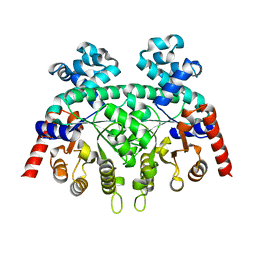 | | Crystal Structure of Plasmodium Berghei Orotidine 5'-monophosphate Decarboxylase (ortholog of Plasmodium falciparum PF10_0225) | | Descriptor: | IODIDE ION, orotidine-monophosphate-decarboxylase | | Authors: | Qiu, W, Dong, A, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Alam, Z, Melone, M, Weigelt, J, Sundstrom, M, Edwards, A, Arrowsmith, C, Hui, R, Gao, M, Bochkarev, A, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1HCZ
 
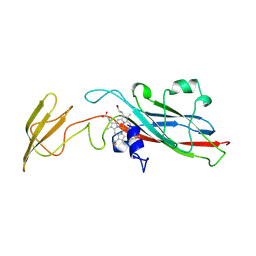 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
1CTM
 
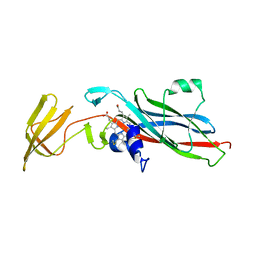 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1TKW
 
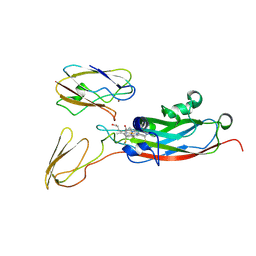 | | The transient complex of poplar plastocyanin with turnip cytochrome f determined with paramagnetic NMR | | Descriptor: | COPPER (II) ION, Cytochrome f, HEME C, ... | | Authors: | Lange, C, Cornvik, T, Diaz-Moreno, I, Ubbink, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The transient complex of poplar plastocyanin with cytochrome f: effects of ionic strength and pH
Biochim.Biophys.Acta, 1707, 2005
|
|
2PCF
 
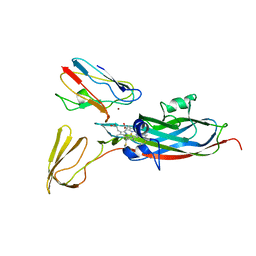 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | Authors: | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
2V1W
 
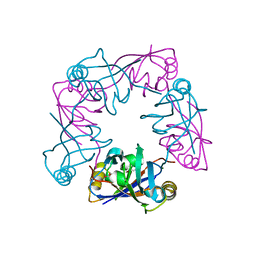 | | Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PDZ AND LIM DOMAIN PROTEIN 4, ... | | Authors: | Soundararajan, M, Shrestha, L, Pike, A.C.W, Salah, E, Burgess-Brown, N, Elkins, J, Umeano, C, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
4DQM
 
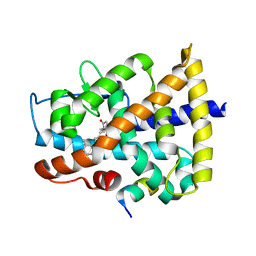 | | Revealing a marine natural product as a novel agonist for retinoic acid receptors with a unique binding mode and antitumor activity | | Descriptor: | (5S)-4-[(3E,7E)-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,7-dien-1-yl]-5-hydroxyfuran-2(5H)-one, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Wang, S, Wang, Z, Lin, S, Zheng, W, Wang, R, Jin, S, Chen, J, Jin, L, Li, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells.
Biochem.J., 446, 2012
|
|
7WQQ
 
 | | Retinoic acid receptor alpha mutant - N299H | | Descriptor: | 4-[(E)-3-(3,5-ditert-butylphenyl)-3-oxidanylidene-prop-1-enyl]benzoic acid, Peptide from Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Huang, X.X, Ng, L.M, Teh, B.T. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effects of breast fibroepithelial tumor associated retinoic acid receptor alpha ligand binding domain mutations on receptor function and retinoid signaling
To Be Published
|
|
6XWG
 
 | | Crystal Structure of the Human RXR/RAR DNA-Binding Domain Heterodimer Bound to the Human RARb2 DR5 Response Element | | Descriptor: | CHLORIDE ION, GLYCEROL, RARb2 DR5 Response Element, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Peluso-Iltis, C, Rochel, N. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR.
Nucleic Acids Res., 48, 2020
|
|
5K13
 
 | | Crystal structure of the RAR alpha ligand-binding domain in complex with an antagonist | | Descriptor: | 4-{5-(3-tert-butylphenyl)-1-[4-(methylsulfonyl)phenyl]-1H-pyrazol-3-yl}benzoic acid, Retinoic acid receptor alpha | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of potent and selective retinoic acid receptor gamma (RAR gamma ) antagonists for the treatment of osteoarthritis pain using structure based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3A9E
 
 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
1DSZ
 
 | | STRUCTURE OF THE RXR/RAR DNA-BINDING DOMAIN HETERODIMER IN COMPLEX WITH THE RETINOIC ACID RESPONSE ELEMENT DR1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*G)-3'), RETINOIC ACID RECEPTOR ALPHA, ... | | Authors: | Rastinejad, F, Wagner, T, Zhao, Q, Khorasanizadeh, S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1.
EMBO J., 19, 2000
|
|
5YUF
 
 | | Crystal Structure of PML RING tetramer | | Descriptor: | Protein PML, ZINC ION | | Authors: | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
1IJK
 
 | | The von Willebrand Factor mutant (I546V) A1 domain-botrocetin Complex | | Descriptor: | Botrocetin, von Willebrand factor | | Authors: | Fukuda, K, Doggett, T.A, Bankston, L.A, Cruz, M.A, Diacovo, T.G, Liddington, R.C. | | Deposit date: | 2001-04-26 | | Release date: | 2002-07-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of von Willebrand factor activation by the snake toxin botrocetin.
Structure, 10, 2002
|
|
3IQ3
 
 | | Crystal Structure of Bothropstoxin-I complexed with polietilene glicol 4000 - crystallized at 283 K | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Phospholipase A2 homolog bothropstoxin-1, SULFATE ION | | Authors: | Salvador, G.H.M, Marchi-Salvador, D.P, Silva, M.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-08-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I3I
 
 | |
4WTB
 
 | | BthTX-I, a svPLA2s-like toxin, complexed with zinc ions | | Descriptor: | Basic phospholipase A2 homolog bothropstoxin-1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Functional and structural studies of a Phospholipase A2-like protein complexed to zinc ions: Insights on its myotoxicity and inhibition mechanism.
Biochim. Biophys. Acta, 1861, 2017
|
|
3CXI
 
 | | Structure of BthTX-I complexed with alpha-tocopherol | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Myotoxic phospholipase A2-like, SULFATE ION, ... | | Authors: | dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Comparative structural studies on Lys49-phospholipases A(2) from Bothrops genus reveal their myotoxic site.
J.Struct.Biol., 167, 2009
|
|
6DIK
 
 | | Crystal structure of Bothropstoxin I (BthTX-I) complexed to Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, BICARBONATE ION, Basic phospholipase A2 homolog bothropstoxin-1, ... | | Authors: | Cardoso, F.F, Salvador, G.H.M, Borges, R.J. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis of phospholipase A2-like myotoxin inhibition by chicoric acid, a novel potent inhibitor of ophidian toxins.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
1U0N
 
 | |
1U0O
 
 | |
2H8I
 
 | |
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
