5OEN
 
 | | Crystal Structure of STAT2 in complex with IRF9 | | Descriptor: | Interferon regulatory factor 9, Signal transducer and activator of transcription | | Authors: | Rengachari, S, Panne, D. | | Deposit date: | 2017-07-09 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.919 Å) | | Cite: | Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8D3F
 
 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
4E68
 
 | | Unphosphorylated STAT3B core protein binding to dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Collie, G.W, Parkinson, G.N, Shah, R. | | Deposit date: | 2012-03-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography.
Febs Lett., 587, 2013
|
|
7NUF
 
 | | Vaccinia virus protein 018 in complex with STAT1 | | Descriptor: | ACETYL GROUP, SULFATE ION, Signal transducer and activator of transcription 1-alpha/beta, ... | | Authors: | Pantelejevs, T, Talbot-Cooper, C, Smith, G.L, Hyvonen, M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.0004015 Å) | | Cite: | Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Cell Host Microbe, 30, 2022
|
|
6TLC
 
 | | Unphosphorylated human STAT3 in complex with MS3-6 monobody | | Descriptor: | Monobody, Signal transducer and activator of transcription 3 | | Authors: | La Sala, G, Lau, K, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Selective inhibition of STAT3 signaling using monobodies targeting the coiled-coil and N-terminal domains.
Nat Commun, 11, 2020
|
|
6QHD
 
 | | Lysine acetylated and tyrosine phosphorylated STAT3 in a complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Arbely, E, Belo, Y, Shahar, A, Zarivach, R. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
3CWG
 
 | | Unphosphorylated mouse STAT3 core fragment | | Descriptor: | Signal transducer and activator of transcription 3 | | Authors: | Ren, Z, Mao, X, Mertens, C, Krishnaraj, R, Qin, J, Mandal, P.K, Romanowshi, M.J, McMurray, J.S. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of unphosphorylated STAT3 core fragment.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
5D39
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
1BG1
 
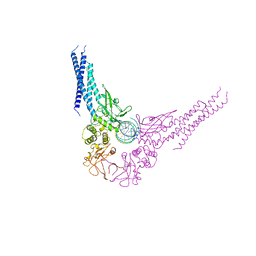 | | TRANSCRIPTION FACTOR STAT3B/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR STAT3B) | | Authors: | Becker, S, Groner, B, Muller, C.W. | | Deposit date: | 1998-06-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Three-dimensional structure of the Stat3beta homodimer bound to DNA.
Nature, 394, 1998
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
4Y5U
 
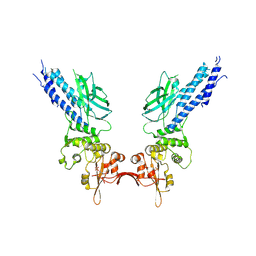 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8T12
 
 | |
8T13
 
 | |
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
1Y1U
 
 | | Structure of unphosphorylated STAT5a | | Descriptor: | Signal transducer and activator of transcription 5A | | Authors: | Neculai, D, Neculai, A.M, Verrier, S, Straub, K, Klumpp, K, Pfitzner, E, Becker, S. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure of the unphosphorylated STAT5a dimer
J.Biol.Chem., 280, 2005
|
|
6MBZ
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural and functional consequences of the STAT5BN642Hdriver mutation.
Nat Commun, 10, 2019
|
|
6MBW
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural and functional consequences of the STAT5BN642H driver mutation.
Nat Commun, 10, 2019
|
|
1YVL
 
 | | Structure of Unphosphorylated STAT1 | | Descriptor: | 5-residue peptide, GOLD ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Mao, X, Ren, Z, Parker, G.N, Sondermann, H, Pastorello, M.A, Wang, W, McMurray, J.S, Demeler, B, Darnell Jr, J.E, Chen, X. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of unphosphorylated STAT1 association and receptor binding.
Mol.Cell, 17, 2005
|
|
8YYV
 
 | | A dimeric STAT1-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*TP*G)-3'), Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Sugiyama, A, Minami, M, Sugita, Y, Ose, T. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein.
Sci.Signal., 18, 2025
|
|
