5LYO
 
 | |
8EDV
 
 | |
6F0E
 
 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
4IC0
 
 | | Crystal Structure of PAI-1 in Complex with Gallate | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Plasminogen activator inhibitor 1 | | Authors: | Hong, Z.B, Lin, Z.H, Gong, L.H, Huang, M.D. | | Deposit date: | 2012-12-09 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of PAI-1 in Complex with Gallate
To be Published
|
|
5EZD
 
 | | Crystal structure of a Hepatocyte growth factor activator inhibitor-1 (HAI-1) fragment covering the PKD-like 'internal' domain and Kunitz domain 1 | | Descriptor: | ACETATE ION, Kunitz-type protease inhibitor 1, POTASSIUM ION | | Authors: | Hong, Z, Andreasen, P.A, Morth, J.P, Jensen, J.K. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Two-domain Fragment of Hepatocyte Growth Factor Activator Inhibitor-1: FUNCTIONAL INTERACTIONS BETWEEN THE KUNITZ-TYPE INHIBITOR DOMAIN-1 AND THE NEIGHBORING POLYCYSTIC KIDNEY DISEASE-LIKE DOMAIN.
J.Biol.Chem., 291, 2016
|
|
2MSX
 
 | | The solution structure of the MANEC-type domain from Hepatocyte Growth Factor Inhibitor 1 reveals an unexpected PAN/apple domain-type fold | | Descriptor: | Kunitz-type protease inhibitor 1 | | Authors: | Hong, Z, Nowakowski, M.E, Spronk, C, Petersen, S.V, Petersen, J.S, Kozminski, W, Mulder, F, Jensen, J.K. | | Deposit date: | 2014-08-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Biochem.J., 466, 2015
|
|
5Y7N
 
 | | Crystal structure of AKR1B10 in complex with NADP+ and Androst-4-ene-3-beta-6-alpha-diol | | Descriptor: | (3S,6S,8S,9S,10R,13S,14R)-10,13-dimethyl-2,3,6,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,6-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hong, Z, Liping, Z, Cuiyun, L, Wei, Z. | | Deposit date: | 2017-08-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants for the inhibition of AKR1B10 by steroids
To Be Published
|
|
7ZGB
 
 | | Structure of yeast Sec14p with NPPM112 | | Descriptor: | 4-fluoranyl-~{N}-[(4-pyrrolidin-1-ylphenyl)methyl]benzamide, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGD
 
 | | Structure of yeast Sec14p with NPPM244 | | Descriptor: | (4-bromanyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGC
 
 | | Structure of yeast Sec14p with NPPM481 | | Descriptor: | (4-chloranyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, PHOSPHATE ION, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGA
 
 | | Structure of yeast Sec14p with ergoline | | Descriptor: | SEC14 cytosolic factor, ~{O}9-methyl ~{O}4-[2,2,2-tris(chloranyl)ethyl] (5~{a}~{S},6~{a}~{S},9~{R},10~{a}~{S})-7-methyl-3-nitro-5,5~{a},6,6~{a},8,9,10,10~{a}-octahydroindolo[4,3-fg]quinoline-4,9-dicarboxylate | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZG9
 
 | | Structure of yeast Sec14p with himbacine | | Descriptor: | Himbacine, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
1C2P
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VIP
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008 | | Descriptor: | TP53-binding protein 1, UNKNOWN ATOM OR ION, {4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]phenyl}(4-ethylpiperazin-1-yl)methanone | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008
to be published
|
|
8EOM
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
1TP7
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
1XRJ
 
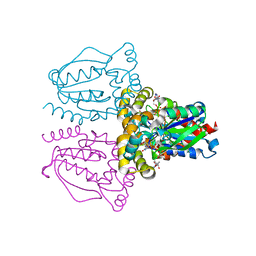 | | Rapid structure determination of human uridine-cytidine kinase 2 using a conventional laboratory X-ray source and a single samarium derivative | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Appleby, T.C, Larson, G, Wu, J.Z, Cheney, I.W, Hong, Z, Yao, N. | | Deposit date: | 2004-10-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human uridine-cytidine kinase 2 determined by SIRAS using a rotating-anode X-ray generator and a single samarium derivative.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4XSK
 
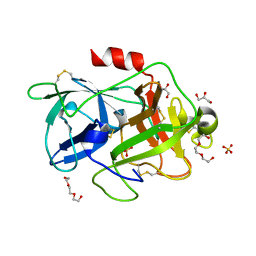 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
