8DS5
 
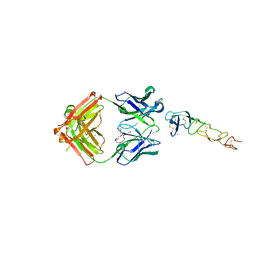 | | X-ray structure of the MK5890 Fab - CD27 antibody-antigen complex | | Descriptor: | CADMIUM ION, CD27 antigen, MK-5890 Fab heavy chain, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Preclinical characterization and clinical translation of pharmacodynamic markers for MK-5890: a human CD27 activating antibody for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
6MUL
 
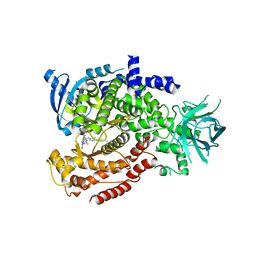 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
6MUM
 
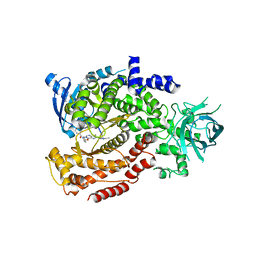 | | Murine PI3K delta kinsae domain - cpd 3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, [(3S)-3-{[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]oxy}pyrrolidin-1-yl](oxan-4-yl)methanone | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
5KX8
 
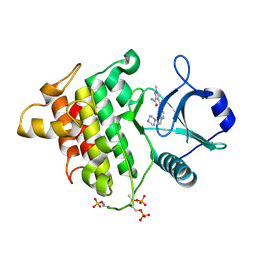 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-5-piperazin-1-yl-pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KX9
 
 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6P)-2-{(3S)-1-[(2-methoxypyrimidin-5-yl)methyl]piperidin-3-yl}-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomic resolution mechanistic studies of ribocil: A highly selective unnatural ligand mimic of the E. coli FMN riboswitch.
Rna Biol., 13, 2016
|
|
5KX7
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-6-(1-methylpyrazol-4-yl)pyridine-2-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3U9N
 
 | | X-ray crystal structure of compound 1 bound to human CHK1 kinase domain | | Descriptor: | 2-(2,3-dihydro-1-benzofuran-5-yl)-N-[2-(piperazin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Novel Series of CHK1 Kinase Inhibitors with Distinctive Hinge Binding Mode
ACS MED.CHEM.LETT., 2012
|
|
6X3N
 
 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5T1S
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1T
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8FPI
 
 | | Co-structure of the Respiratory Syncytial Virus RNA-dependent RNA polymerase with MRK-1 | | Descriptor: | 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6-(4-fluorophenyl)pyridine-2-carbohydrazide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Fischmann, T.O. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Conserved allosteric inhibitory site on the respiratory syncytial virus and human metapneumovirus RNA-dependent RNA polymerases.
Commun Biol, 6, 2023
|
|
8FPJ
 
 | | Co-structure of the Human Metapneunomovirus RNA-dependent RNA polymerase with MRK-1 | | Descriptor: | 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6-(4-fluorophenyl)pyridine-2-carbohydrazide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Fischmann, T.O. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Conserved allosteric inhibitory site on the respiratory syncytial virus and human metapneumovirus RNA-dependent RNA polymerases.
Commun Biol, 6, 2023
|
|
6X3O
 
 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5ENZ
 
 | | S. aureus MnaA-UDP co-structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemical Genetic Analysis and Functional Characterization of Staphylococcal Wall Teichoic Acid 2-Epimerases Reveals Unconventional Antibiotic Drug Targets.
Plos Pathog., 12, 2016
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1FDL
 
 | |
7UYD
 
 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7SSB
 
 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
7UYA
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
5C45
 
 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6M)-2-[(3S)-1-{[2-(methylamino)pyrimidin-5-yl]methyl}piperidin-3-yl]-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Selective small-molecule inhibition of an RNA structural element.
Nature, 526, 2015
|
|
5FBO
 
 | | BTK-inhibitor co-structure | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R},6~{S})-1-cyclopropylcarbonyl-6-methyl-piperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-3-fluoranyl-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
ACS Med Chem Lett, 7, 2016
|
|
4XS2
 
 | | Irak4-inhibitor co-structure | | Descriptor: | (1R,2S,3R,5R)-3-({5-(1,3-benzothiazol-2-yl)-6-chloro-2-[(3-methoxypropyl)amino]pyrimidin-4-yl}amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and hit-to-lead optimization of 2,6-diaminopyrimidine inhibitors of interleukin-1 receptor-associated kinase 4.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YO6
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation.
Acs Med.Chem.Lett., 6, 2015
|
|
