1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4TX4
 
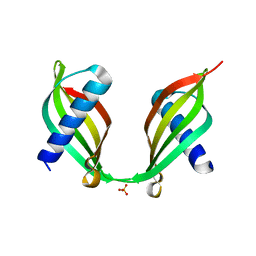 | | Crystal Structure of a Single-Domain Cysteine Protease Inhibitor from Cowpea (Vigna unguiculata) | | Descriptor: | Cysteine proteinase inhibitor, SULFATE ION | | Authors: | Pereira, H.M, Valadares, N, Monteiro-Junior, J.E, Carvalho, C.P.S, Grangeiro, T.B. | | Deposit date: | 2014-07-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression in Escherichia coli of cysteine protease inhibitors from cowpea (Vigna unguiculata): The crystal structure of a single-domain cystatin gives insights on its thermal and pH stability.
Int. J. Biol. Macromol., 102, 2017
|
|
2HLD
 
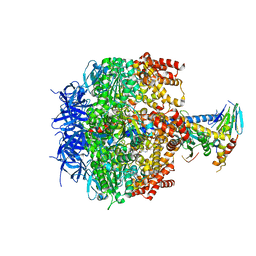 | | Crystal structure of yeast mitochondrial F1-ATPase | | Descriptor: | ATP synthase alpha chain, mitochondrial, ATP synthase beta chain, ... | | Authors: | Kabaleeswaran, V, Puri, N, Walker, J.E, Leslie, A.G, Mueller, D.M. | | Deposit date: | 2006-07-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Embo J., 25, 2006
|
|
2ZFC
 
 | | X-ray crystal structure of an engineered N-terminal HIV-1 GP41 trimer with enhanced stability and potency | | Descriptor: | HIV-1 GP41 | | Authors: | Dwyer, J.J, Wilson, K.L, Martin, K, Seedorff, J.E, Hasan, A, Kim, H. | | Deposit date: | 2007-12-29 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency
Protein Sci., 17, 2008
|
|
2Z85
 
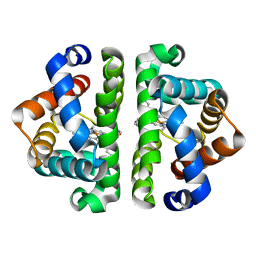 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: M37F Unliganded | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
1EBH
 
 | |
8CDM
 
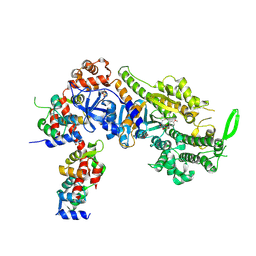 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
4I6R
 
 | |
4IA8
 
 | |
4I6U
 
 | | Crystal Structure of a Y37F mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-11-30 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Mutagenic Analysis of the RM Controller Protein C.Esp1396I.
Plos One, 9, 2014
|
|
5FZN
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SULFATE ION, benzenesulfonamide | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNR
 
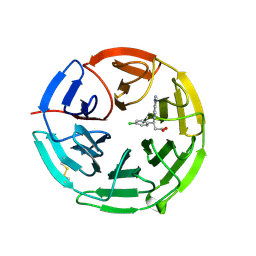 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNS
 
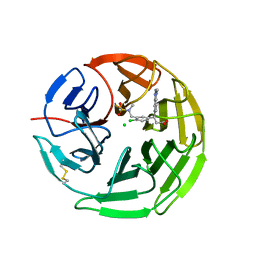 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
4I8T
 
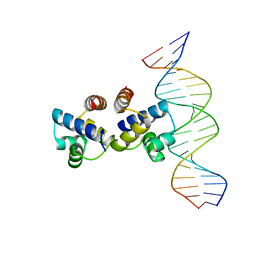 | | C.Esp1396I bound to a 19 base pair DNA duplex | | Descriptor: | DNA (5'-D(P*TP*GP*TP*GP*TP*GP*AP*TP*TP*AP*TP*AP*GP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*GP*AP*CP*TP*AP*TP*AP*AP*TP*CP*AP*CP*AP*CP*A)-3'), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-12-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5FNQ
 
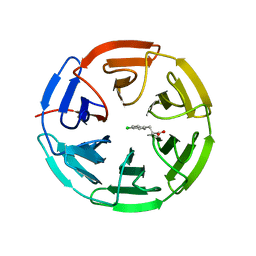 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
4I6T
 
 | |
2VCM
 
 | | Isopenicillin N synthase with substrate analogue AsMCOV | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
2VAU
 
 | | Isopenicillin N synthase with substrate analogue ACOMP (unexposed) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N6^-[(1R)-2-[(1S)-1-CARBOXY-2-(METHYLSULFANYL)ETHOXY]-2-OXO-1-(SULFANYLMETHYL)ETHYL]-6-OXO-L-LYSINE | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-04 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isopenicillin N Synthase Mediates Thiolate Oxidation to Sulfenate in a Depsipeptide Substrate Analogue: Implications for Oxygen Binding and a Link to Nitrile Hydratase?
J.Am.Chem.Soc., 130, 2008
|
|
2VE1
 
 | | Isopenicillin N synthase with substrate analogue AsMCOV (oxygen exposed 1min 20bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
2VBB
 
 | | Isopenicillin N synthase with substrate analogue ACOMP (35minutes oxygen exposure) | | Descriptor: | FE (II) ION, GLYCEROL, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Isopenicillin N Synthase Mediates Thiolate Oxidation to Sulfenate in a Depsipeptide Substrate Analogue: Implications for Oxygen Binding and a Link to Nitrile Hydratase?
J.Am.Chem.Soc., 130, 2008
|
|
2VBD
 
 | | Isopenicillin N synthase with substrate analogue L,L,L-ACOMP (unexposed) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R)-2-[(1R)-1-carboxy-2-(methylsulfanyl)ethoxy]-2-oxo-1-(sulfanylmethyl)ethyl]-6-oxo-L-lysine | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Lll-Configured Depsipeptide Substrate Analogue Bound to Isopenicillin N Synthase.
Org.Biomol.Chem., 8, 2010
|
|
2VBP
 
 | | Isopenicillin N synthase with substrate analogue L,L,L-ACAB (unexposed) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R)-2-{[(1S)-1-carboxypropyl]amino}-2-oxo-1-(sulfanylmethyl)ethyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Studies on the Binding of Selectively Deuterated Lld- and Lll-Substrate Epimers by Isopenicillin N Synthase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
