6Y16
 
 | | CRYSTAL STRUCTURE OF TMARGBP DOMAIN 1 IN COMPLEX WITH THE GUANIDINIUM ION | | Descriptor: | 1,2-ETHANEDIOL, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Ruggiero, A, Balasco, N, Smaldone, G, Graziano, G, Vitagliano, L. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium binding to proteins: The intriguing effects on the D1 and D2 domains of Thermotoga maritima Arginine Binding Protein and a comprehensive analysis of the Protein Data Bank.
Int.J.Biol.Macromol., 163, 2020
|
|
6NJL
 
 | |
5L4E
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with thiopental | | Descriptor: | 5-ethyl-5-[(2R)-pentan-2-yl]-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, CHLORIDE ION, DODECANE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
8SPN
 
 | | Crystal structure of macrophage migration inhibitory factor in complex with T614 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Iguratimod, ... | | Authors: | Pantouris, G, Lolis, E. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Iguratimod, an allosteric inhibitor of macrophage migration inhibitory factor (MIF), prevents mortality and oxidative stress in a murine model of acetaminophen overdose.
Mol Med, 30, 2024
|
|
1A37
 
 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
5L4H
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with 5-(2-BROMO-ETHYL)-5-ETHYL-PYRIMIDINE-2,4,6-TRIONE (brominated barbiturate) | | Descriptor: | 5-(2-bromoethyl)-5-ethyl-1,3-diazinane-2,4,6-trione, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2017-02-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
2DYV
 
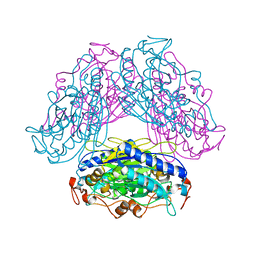 | |
6Z9X
 
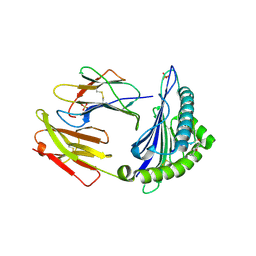 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
1ZCN
 
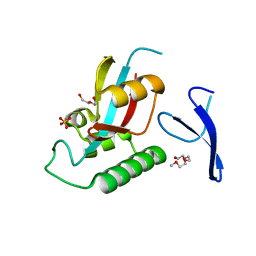 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4HHF
 
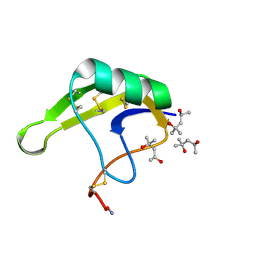 | | Crystal Structure of chemically synthesized scorpion alpha-toxin OD1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-toxin OD1 | | Authors: | Wang, C.I.A, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-10-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical engineering and structural and pharmacological characterization of the alpha-scorpion toxin OD1.
Acs Chem.Biol., 8, 2013
|
|
6HY7
 
 | |
8T1I
 
 | | Atomic model of the mammalian Mediator complex with MED26 subunit | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
1B57
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCOLOHYDROXAMIC ACID, PROTEIN (FRUCTOSE-BISPHOSPHATE ALDOLASE II), ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity.
J.Mol.Biol., 287, 1999
|
|
8T9D
 
 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
5L47
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with cyanoselenobarbital (seleniated barbiturate) | | Descriptor: | 2-[5-ethyl-2,4,6-tris(oxidanylidene)-1,3-diazinan-5-yl]ethyl selenocyanate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Reinholds Ruza, R, Fourati, Z, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
1LQ2
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
7DCA
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
2ECU
 
 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3QPY
 
 | | Crystal structure of a mutant (K57A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
8ARN
 
 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with an endogenous tetrapeptide | | Descriptor: | Endogenous tetrapeptide (SER-ASN-SER-SER), Oligopeptide-binding protein OppA | | Authors: | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
1L7H
 
 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
3GS7
 
 | | Human transthyretin (TTR) complexed with (E)-3-(2-methoxybenzylideneaminooxy)propanoic acid (inhibitor 13) | | Descriptor: | 3-({[(1Z)-(2-methoxyphenyl)methylidene]amino}oxy)propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
