7ET3
 
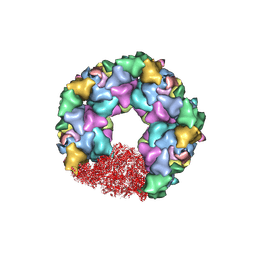 | | C5 portal vertex in the enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
3MGC
 
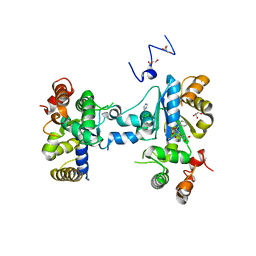 | | Teg12 Apo | | Descriptor: | GLYCEROL, IMIDAZOLE, N-L-ALPHA-ASPARTYL L-PHENYLALANINE 1-METHYL ESTER, ... | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of the glycopeptide sulfotransferase Teg12 in a complex with the teicoplanin aglycone.
Biochemistry, 49, 2010
|
|
7YQ3
 
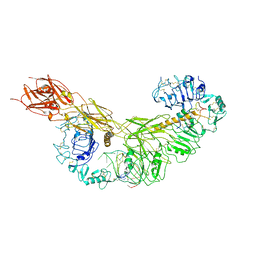 | | human insulin receptor bound with A43 DNA aptamer and insulin | | Descriptor: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ4
 
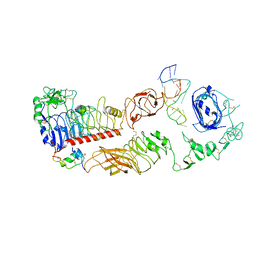 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ6
 
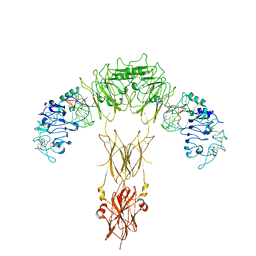 | | human insulin receptor bound with A62 DNA aptamer | | Descriptor: | IR-A62 aptamer, Isoform Short of Insulin receptor | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
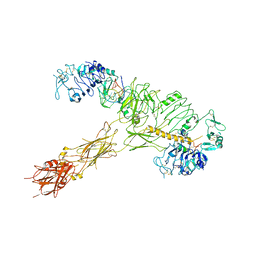 | | human insulin receptor bound with A62 DNA aptamer and insulin | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
6MY0
 
 | |
6N65
 
 | | KRAS G-quadruplex G16T mutant. | | Descriptor: | KRAS G-quadruplex G16T mutant, POTASSIUM ION | | Authors: | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | Deposit date: | 2018-11-25 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
7D5D
 
 | | Left-handed G-quadruplex containing one bulge | | Descriptor: | 1xBulge-LHG4motif, POTASSIUM ION, SPERMINE | | Authors: | Das, P, Ngo, K.H, Winnerdy, F.R, Maity, A, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
7D5E
 
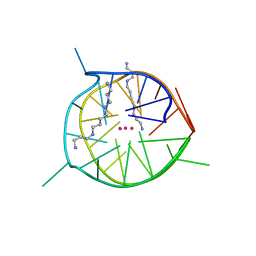 | | Left-handed G-quadruplex containing two bulges | | Descriptor: | 2xBulge-LHG4motif, POTASSIUM ION, SODIUM ION, ... | | Authors: | Das, P, Maity, A, Ngo, K.H, Winnerdy, F.R, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
3OPT
 
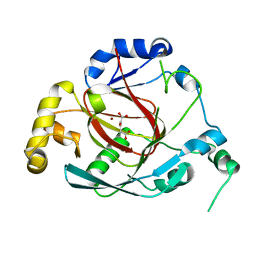 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3N5E
 
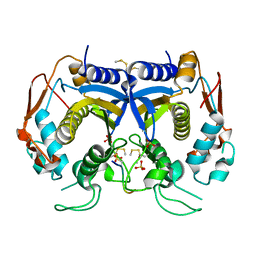 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7BI0
 
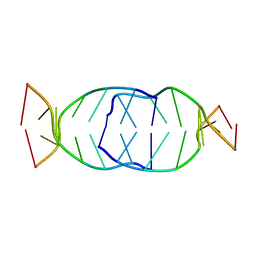 | | GA repetition with i-motif clip at 5'-end | | Descriptor: | (CH+)C(CH+)GAGA, C(CH+)CGAGA | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
7BL0
 
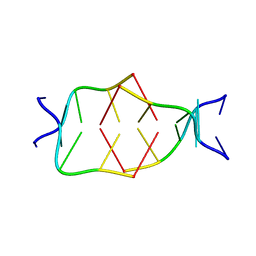 | | GA attached to an i-motif clip at 3'-end | | Descriptor: | GA(CH+)C(5MeCH+), GAC(HCY)(5MeC) | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
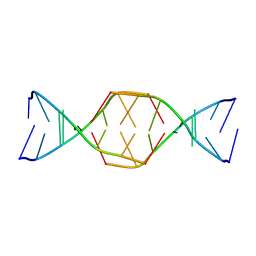
7BLM
 
 | |
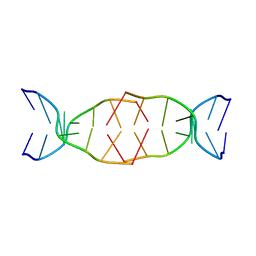
7BMA
 
 | |
3NLB
 
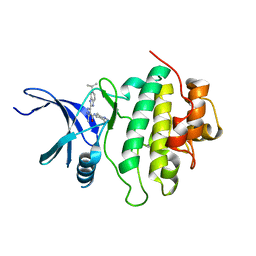 | | Novel kinase profile highlights the temporal basis of context dependent checkpoint pathways to cell death | | Descriptor: | 3-methyl-5-[5-(1-methylethyl)-1H-benzimidazol-2-yl]-N-(1-methylpiperidin-4-yl)-1H-pyrazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Massey, A.J, Borgognoni, J, Bentley, C, Foloppe, N, Fiumana, A, Walmsley, L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent cell cycle checkpoint abrogation by a novel kinase inhibitor
Plos One, 5, 2010
|
|
3P9N
 
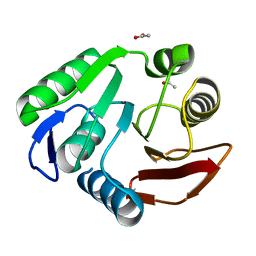 | | Rv2966c of M. tuberculosis is a RsmD-like methyltransferase | | Descriptor: | ACETATE ION, POSSIBLE METHYLTRANSFERASE (METHYLASE) | | Authors: | Kumar, A, Malhotra, K, Saigal, K, Sinha, K.M, Taneja, B. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of Rv2966c protein reveals an RsmD-like methyltransferase from Mycobacterium tuberculosis and the role of its N-terminal domain in target recognition
J.Biol.Chem., 286, 2011
|
|
1XPU
 
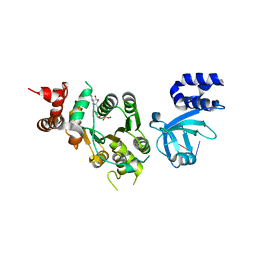 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1XPO
 
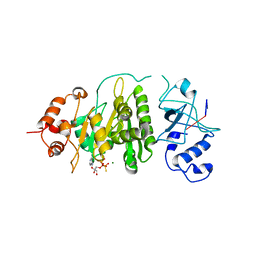 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
7YIB
 
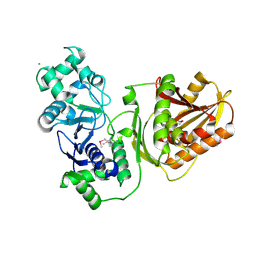 | | Crystal structure of wild-type Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, CHLORIDE ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
7YIA
 
 | | Crystal structure of K74A mutant of Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, MAGNESIUM ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
7D5F
 
 | | Left-handed G-quadruplex containing 3 bulges | | Descriptor: | 3xBulge-LHG4motif | | Authors: | Winnerdy, F.R, Das, P, Ngo, K.H, Maity, A, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
7WKV
 
 | | Crystal structure of human ALKBH5 in complex with 2-oxoglutarate (2OG) and m6A-containing ssRNA | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
7WL0
 
 | | Crystal structure of human ALKBH5 in complex with N-oxalylglycine (NOG) and m6A-containing ssRNA | | Descriptor: | FORMIC ACID, MANGANESE (II) ION, N-OXALYLGLYCINE, ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
