1CDH
 
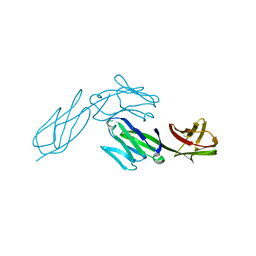 | |
1ATU
 
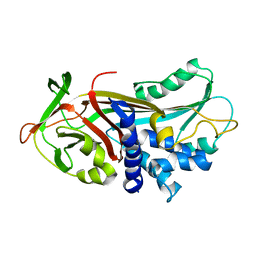 | | UNCLEAVED ALPHA-1-ANTITRYPSIN | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Ryu, S.-E, Choi, H.-J. | | Deposit date: | 1997-05-11 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The native strains in the hydrophobic core and flexible reactive loop of a serine protease inhibitor: crystal structure of an uncleaved alpha1-antitrypsin at 2.7 A.
Structure, 4, 1996
|
|
1CDI
 
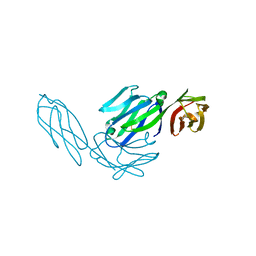 | |
3S4E
 
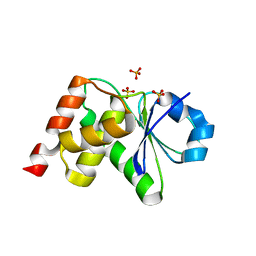 | | Crystal Structrue of a Novel Mitogen-activated Protein Kinase Phosphatase, SKRP1 | | Descriptor: | Dual specificity protein phosphatase 19, PHOSPHATE ION, SULFATE ION | | Authors: | Wei, C.H, Ryu, S.Y, Jeon, Y.H, Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2011-05-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of a novel mitogen-activated protein kinase phosphatase, SKRP1.
Proteins, 79, 2011
|
|
8GUY
 
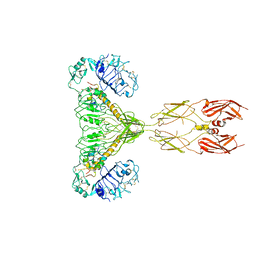 | | human insulin receptor bound with two insulin molecules | | Descriptor: | Insulin A chain, Insulin, isoform 2, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
8H5A
 
 | |
8H58
 
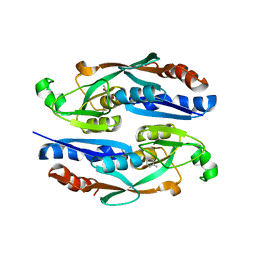
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
3M7M
 
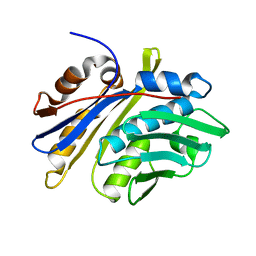 | | Crystal structure of monomeric hsp33 | | Descriptor: | 33 kDa chaperonin | | Authors: | Chi, S.W, Jeong, D.G, Woo, J.R, Park, B.C, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-03-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of monomeric hsp33
To be Published
|
|
4JNB
 
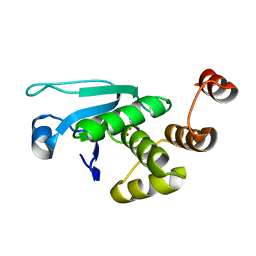 | | Crystal structure of the Catalytic Domain of Human DUSP12 | | Descriptor: | Dual specificity protein phosphatase 12, SULFATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JMJ
 
 | | Structure of dusp11 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JMK
 
 | | Structure of dusp8 | | Descriptor: | Dual specificity protein phosphatase 8, SULFATE ION | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KI9
 
 | | Crystal structure of the catalytic domain of human DUSP12 at 2.0 A resolution | | Descriptor: | Dual specificity protein phosphatase 12, PHOSPHATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-05-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1PRX
 
 | | HORF6 A NOVEL HUMAN PEROXIDASE ENZYME | | Descriptor: | HORF6 | | Authors: | Choi, H.-J, Kang, S.W, Yang, C.-H, Rhee, S.G, Ryu, S.-E. | | Deposit date: | 1998-04-03 | | Release date: | 1998-06-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
1HP7
 
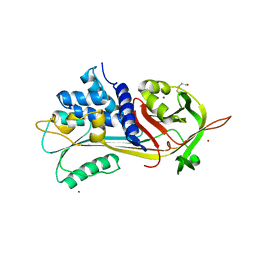 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | Descriptor: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | Authors: | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
5D4Z
 
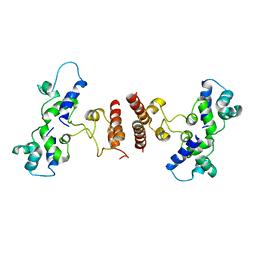 | |
5D50
 
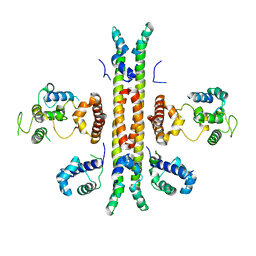 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1I7F
 
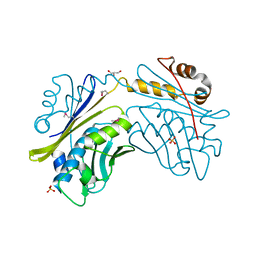 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | Authors: | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-05-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
1KU0
 
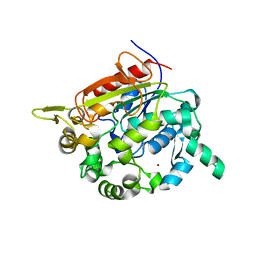 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
1WOU
 
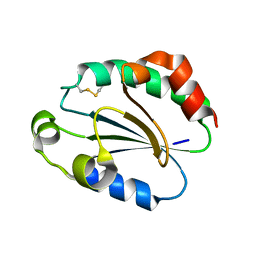 | | Crystal Structure of human Trp14 | | Descriptor: | thioredoxin -related protein, 14 kDa | | Authors: | Woo, J.R, Kim, S.J, Jeong, W, Cho, Y.H, Lee, S.C, Chung, Y.J, Rhee, S.G, Ryu, S.E. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of cellular redox regulation by human TRP14
J.Biol.Chem., 279, 2004
|
|
1XM2
 
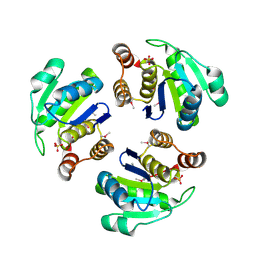 | | Crystal structure of Human PRL-1 | | Descriptor: | SULFATE ION, Tyrosine Phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Kim, J.H, Son, J.H, Ryu, S.E. | | Deposit date: | 2004-10-01 | | Release date: | 2005-01-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms
J.Mol.Biol., 345, 2005
|
|
2EH7
 
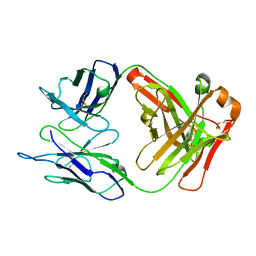 | | Crystal structure of humanized KR127 FAB | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EH8
 
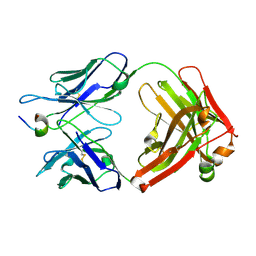 | | Crystal structure of the complex of humanized KR127 fab and PRES1 peptide epitope | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1I69
 
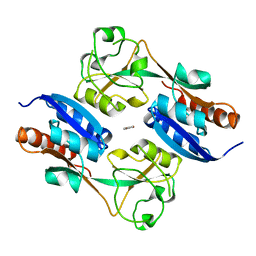 | | CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR | | Descriptor: | BENZOIC ACID, HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1NM3
 
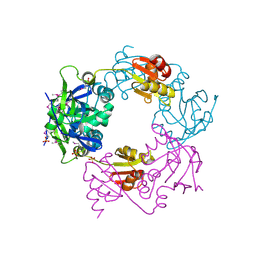 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | Descriptor: | Protein HI0572, SULFATE ION | | Authors: | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
1I6A
 
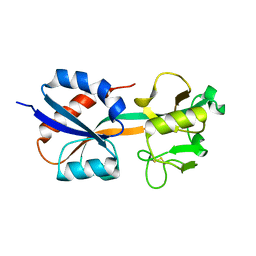 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF OXYR | | Descriptor: | HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
