7JXY
 
 | | Structure of TTBK1 kinase domain in complex with Compound 18 | | Descriptor: | (3S)-1-[1-(2-aminopyrimidin-4-yl)-1H-pyrazolo[4,3-c]pyridin-6-yl]-3-methylpent-1-yn-3-ol, Tau-tubulin kinase 1 | | Authors: | Chodaprambil, J.V. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Tau Tubulin Kinase 1 (TTBK1) Inhibitors that Lower Tau Phosphorylation In Vivo.
J.Med.Chem., 64, 2021
|
|
3VH7
 
 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P21 | | Descriptor: | CP32M, Envelope glycoprotein gp160, MAGNESIUM ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3S0N
 
 | | Crystal Structure of Human Chymase with Benzimidazolone Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{3-[(4-methyl-1-benzothiophen-3-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}butanoic acid, Chymase, ... | | Authors: | Qian, K.C, Farrow, N.A, Padyana, A.K. | | Deposit date: | 2011-05-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Benzimidazolone as potent chymase inhibitor: Modulation of reactive metabolite formation in the hydrophobic (P(1)) region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1P4O
 
 | | Structure of Apo unactivated IGF-1R KInase domain at 1.5A resolution. | | Descriptor: | Insulin-like growth factor I receptor protein | | Authors: | Munshi, S, Kornienko, M, Hall, D.L, Darke, P.L, Waxman, L, Kuo, L.C. | | Deposit date: | 2003-04-23 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4CML
 
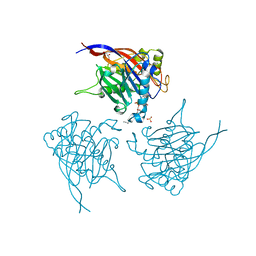 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
6A0K
 
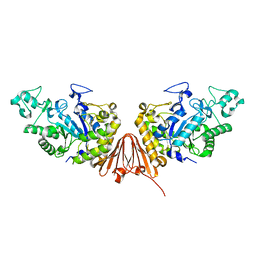 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with panose | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
2F3W
 
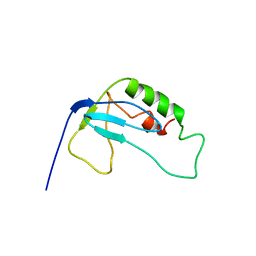 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
4K10
 
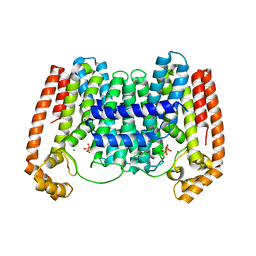 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM and Mg2+ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1TCX
 
 | | HIV TRIPLE MUTANT PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
4COC
 
 | | HIV-1 capsid C-terminal domain mutant (Y169L) | | Descriptor: | CAPSID PROTEIN P24, SULFATE ION | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ARM
 
 | | Crystal Structure of the Complex of Phospholipase A2 with a natural compound atropine at 1.2 A resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2005-08-20 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding
Proteins, 64, 2006
|
|
3R13
 
 | |
2OID
 
 | |
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
3BD7
 
 | | Glycogen Phosphorylase complex with 1(-D-glucopyranosyl) thymine | | Descriptor: | 1-beta-D-glucopyranosyl-5-methylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form, ... | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
4CTO
 
 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(2Z,5R,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-4-oxo-6-oxa-1-thia-3-azaspiro[4.5]dec-2-ylidene]benzamide, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
4OAQ
 
 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
3RMV
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Yue, W.W, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3EL0
 
 | |
4O91
 
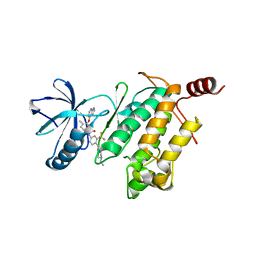 | | Crystal Structure of type II inhibitor NG25 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-4-methyl-3-(1H-pyrrolo[2,3-b]pyridin-4-yloxy)benzamide | | Authors: | Gurbani, D, Hunter, J.C, Tan, L, Westover, K.D. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of Type II Inhibitors of TGF beta-Activated Kinase 1 (TAK1) and Mitogen-Activated Protein Kinase Kinase Kinase Kinase 2 (MAP4K2).
J.Med.Chem., 58, 2015
|
|
4OJR
 
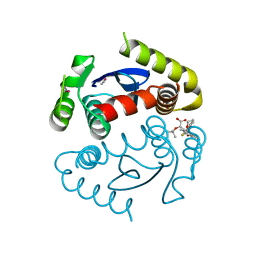 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | Authors: | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | Deposit date: | 2014-01-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
5PZM
 
 | |
3RV2
 
 | |
4D96
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with 1-amino-1-carboxycyclopropane (ACC) | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
2Q9X
 
 | | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1 | | Descriptor: | GLYCEROL, Heparin-binding growth factor 1 | | Authors: | Szlachcic, A, Zakrzewska, M, Krowarsch, D, Os, V, Helland, R, Otlewski, J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1
To be Published
|
|
