6LXM
 
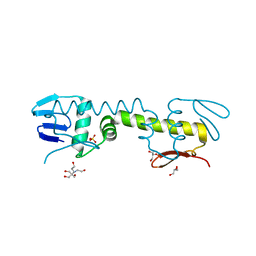 | | Crystal structure of C-terminal DNA-binding domain of Escherichia coli OmpR as a domain-swapped dimer | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Sadotra, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structural basis for promoter DNA recognition by the response regulator OmpR.
J.Struct.Biol., 213, 2020
|
|
6LMK
 
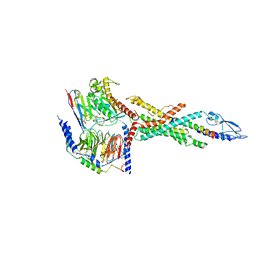 | | Cryo-EM structure of the human glucagon receptor in complex with Gs | | Descriptor: | Glucagon, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A, Han, S, Tai, L, Sun, F, Zhao, Q, Wu, B. | | Deposit date: | 2019-12-26 | | Release date: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of Gsand Girecognition by the human glucagon receptor.
Science, 367, 2020
|
|
8RGT
 
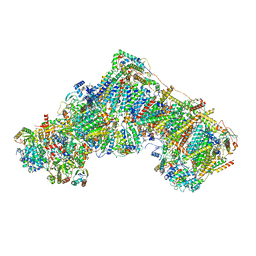 | | Open Complex I from murine brain | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-12-14 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
4IDM
 
 | |
5I3Q
 
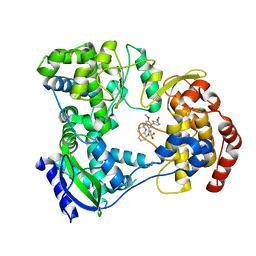 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 29 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5I43
 
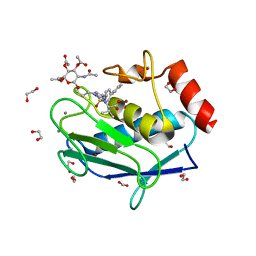 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated triazole-linked carboxylate chelator water-soluble inhibitor (DC32). | | Descriptor: | (2R)-2-[({1-[3-({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}oxy)propyl]-1H-1,2,3-triazol-4-yl}methyl)(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid (non-preferred name), 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
3QXW
 
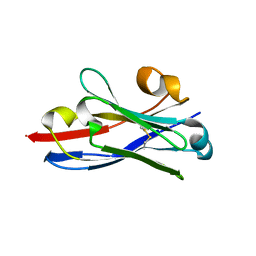 | | Free structure of an anti-methotrexate CDR1-4 Graft VHH Antibody | | Descriptor: | Anti-Methotrexate CDR1-4 Graft VHH, SODIUM ION, SULFATE ION | | Authors: | Fanning, S.W, Horn, J.R. | | Deposit date: | 2011-03-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop.
Protein Sci., 20, 2011
|
|
8RGR
 
 | | Closed Complex I from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-12-14 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
6LPN
 
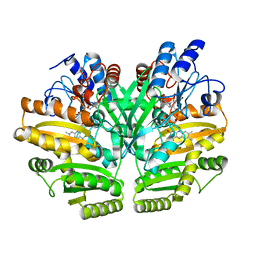 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6M54
 
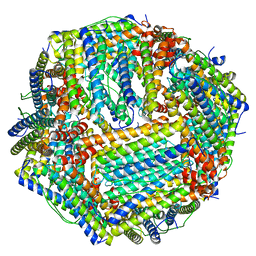 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
4LI1
 
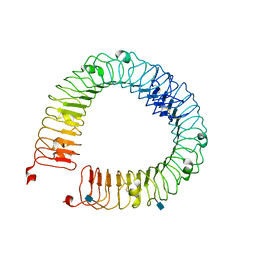 | |
7DOV
 
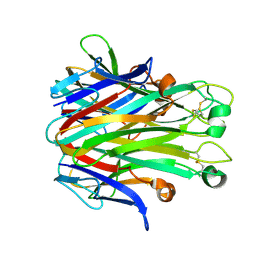 | |
1P16
 
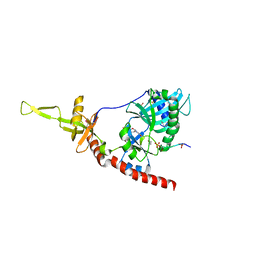 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
3O96
 
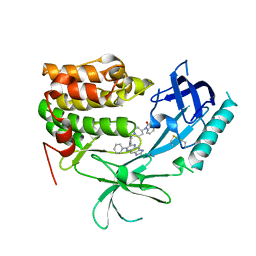 | | Crystal Structure of Human AKT1 with an Allosteric Inhibitor | | Descriptor: | 1-(1-(4-(7-phenyl-1H-imidazo[4,5-g]quinoxalin-6-yl)benzyl)piperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one, RAC-alpha serine/threonine-protein kinase | | Authors: | Voegtli, W.C, Wu, W.-I, Lord-Ondash, H.A, Dizon, F.P, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition.
Plos One, 5, 2010
|
|
6M2I
 
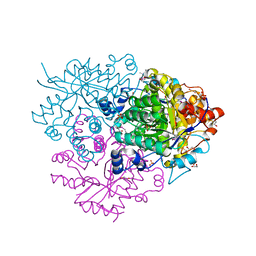 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
4IP1
 
 | |
4IP6
 
 | |
3SS3
 
 | |
3OM0
 
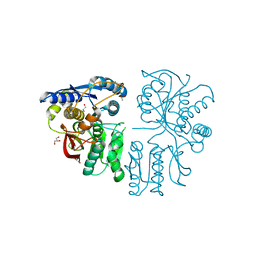 | |
1PCP
 
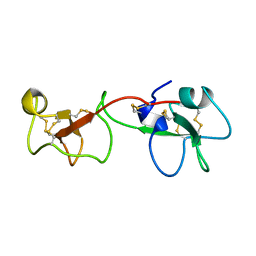 | | SOLUTION STRUCTURE OF A TREFOIL-MOTIF-CONTAINING CELL GROWTH FACTOR, PORCINE SPASMOLYTIC PROTEIN | | Descriptor: | PORCINE SPASMOLYTIC PROTEIN | | Authors: | Carr, M.D, Bauer, C.J, Gradwell, M.J, Feeney, J. | | Deposit date: | 1993-02-04 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a trefoil-motif-containing cell growth factor, porcine spasmolytic protein.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
4IR7
 
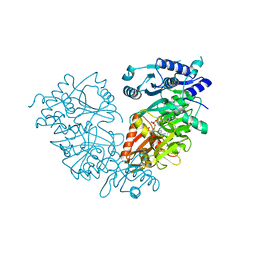 | | Crystal Structure of Mtb FadD10 in Complex with Dodecanoyl-AMP | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, Long chain fatty acid CoA ligase FadD10, MAGNESIUM ION | | Authors: | Liu, Z, Wang, F, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis FadD10 protein reveal a new type of adenylate-forming enzyme.
J.Biol.Chem., 288, 2013
|
|
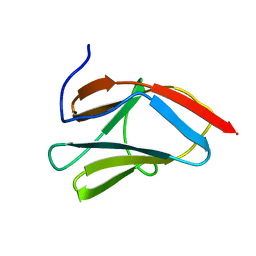
1PD6
 
 | |
4IUY
 
 | | Crystal structure of short-chain dehydrogenase/reductase (apo-form) from A. baumannii clinical strain WM99C | | Descriptor: | Short chain dehydrogenase/reductase | | Authors: | Shah, B.S, Tetu, S.G, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure of a short-chain dehydrogenase/reductase (SDR) within a genomic island from a clinical strain of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3P22
 
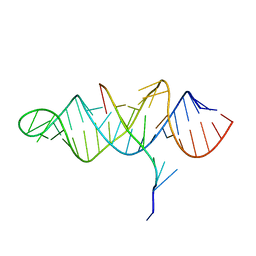 | | Crystal structure of the ENE, a viral RNA stability element, in complex with A9 RNA | | Descriptor: | Core ENE hairpin from KSHV PAN RNA, oligo(A)9 RNA | | Authors: | Mitton-Fry, R.M, DeGregorio, S.J, Wang, J, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Poly(A) tail recognition by a viral RNA element through assembly of a triple helix.
Science, 330, 2010
|
|
4ITL
 
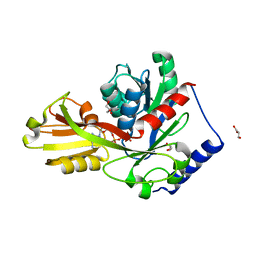 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
