8HCC
 
 | | Crystal structure of mTREX1-RNA product complex (AMP) | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
4WL3
 
 | |
8HCF
 
 | | Crystal structure of mTREX1-UMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Three-prime repair exonuclease 1, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
2VIE
 
 | | Human BACE-1 in complex with N-((1S,2R)-1-benzyl-2-hydroxy-3-((1,1,5- trimethylhexyl)amino)propyl)-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl) benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1,1,5-trimethylhexyl)amino]propyl}-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8HE8
 
 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
4WLN
 
 | | Crystal structure of apo MDH2 | | Descriptor: | Malate dehydrogenase, mitochondrial, PHOSPHATE ION | | Authors: | Eo, Y.M, Han, B.G, Ahn, H.C. | | Deposit date: | 2014-10-07 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of apo MDH2
To Be Published
|
|
2VJ8
 
 | | Complex of human leukotriene A4 hydrolase with a hydroxamic acid inhibitor | | Descriptor: | 6-[{(2S)-2-AMINO-3-[4-(BENZYLOXY)PHENYL]PROPYL}(HYDROXY)AMINO]-6-OXOHEXANOIC ACID), ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Andersson, B, Wong, C.-H, Samuelsson, B, Haeggstrom, J.Z. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Leukotriene A4 Hydrolase in Complex with Captopril and Two Competitive Tight-Binding Inhibitors
Faseb J., 16, 2002
|
|
8HKD
 
 | |
4WM6
 
 | |
2V7A
 
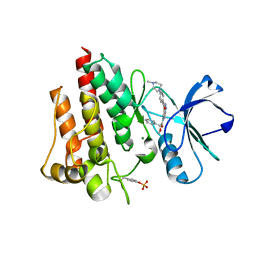 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
8HD3
 
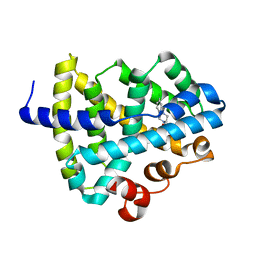 | | Farnesoid X Receptor Agonists_FXR fused with a HD3 peptide | | Descriptor: | Farnesoid X Receptor, [(3s,5s,7s)-adamantan-1-yl][4-(2-amino-5-chlorophenyl)piperazin-1-yl]methanone | | Authors: | Lu, X, Zhang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of FXR fused with a HD3 peptide in complex with agonist QT0127
To Be Published
|
|
2V8L
 
 | |
4WML
 
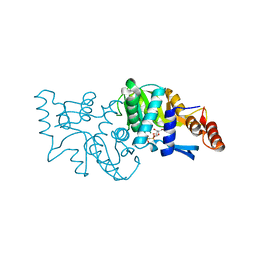 | | Crystal structure of Saccharomyces cerevisiae OMP synthase in complex with PRP(CH2)P | | Descriptor: | 1-O-[(R)-hydroxy(phosphonomethyl)phosphoryl]-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, Orotate phosphoribosyltransferase 1 | | Authors: | Bang, M.B, Molich, U, Hansen, M.R, Grubmeyer, C, Harris, P. | | Deposit date: | 2014-10-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Saccharomyces cerevisia OMP synthase in complex with PRP(CH2)P
To Be Published
|
|
8HEZ
 
 | | Structure of human SGLT2-MAP17 complex with Dapagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4WFR
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
2V8W
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
8HEE
 
 | | Pentamer of FMDV (A/TUR/14/98) | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein | | Authors: | Li, H.Z, Dong, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and in vivo studies of neutralizing antibody topographical classifications reveal mechanisms underlying differences in immunogenicity and antigenicity between 146S and 12S of foot-and-mouth disease virus
To Be Published
|
|
2V93
 
 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | Authors: | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
8HHJ
 
 | |
2VLC
 
 | | Crystal structure of Natural Cinnamomin (Isoform III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein, beta-D-mannopyranose, ... | | Authors: | Azzi, A, Wang, T, Zhu, D.-W, Zou, Y.-S, Liu, W.-Y, Lin, S.-X. | | Deposit date: | 2008-01-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Native Cinnamomin Isoform III and its Comparison with Other Ribosome Inactivating Proteins.
Proteins: Struct., Funct., Bioinf., 74, 2009
|
|
4WEP
 
 | | Apo YehZ from Escerichia coli | | Descriptor: | Putative osmoprotectant uptake system substrate-binding protein OsmF | | Authors: | Kimber, M.S, Lang, S, Mendoza, K, Wood, J.M. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | YehZYXW of Escherichia coli Is a Low-Affinity, Non-Osmoregulatory Betaine-Specific ABC Transporter.
Biochemistry, 54, 2015
|
|
8HDZ
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in an apo form | | Descriptor: | A22 DNA replication processivity factor, E4 uracil-DNA glycosylase, F8 DNA polymerase | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
2VN7
 
 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
4WG1
 
 | | Room temperature crystal structure of lysozyme determined by serial synchrotron crystallography (micro focused beam - crystFEL) | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kapp, U, Shilova, A, Weimhausen, B, Sauter, N.K, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
