1BX2
 
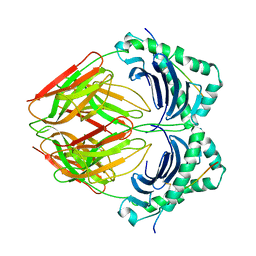 | | CRYSTAL STRUCTURE OF HLA-DR2 (DRA*0101,DRB1*1501) COMPLEXED WITH A PEPTIDE FROM HUMAN MYELIN BASIC PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HLA-DR2) | | Authors: | Smith, K.J, Pyrdol, J, Gauthier, L, Wiley, D.C, Wucherpfennig, K. | | Deposit date: | 1998-10-12 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein.
J.Exp.Med., 188, 1998
|
|
1A1O
 
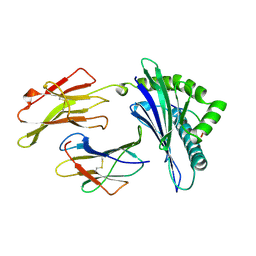 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1A1M
 
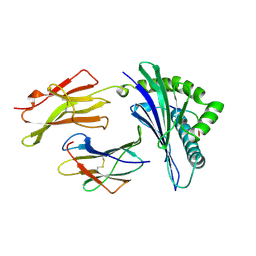 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
5HHG
 
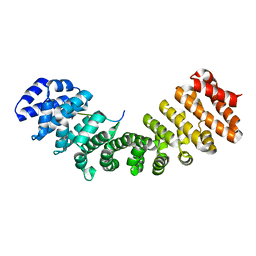 | | Mouse importin alpha: Dengue 2 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, RNA-directed RNA polymerase NS5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
1A1N
 
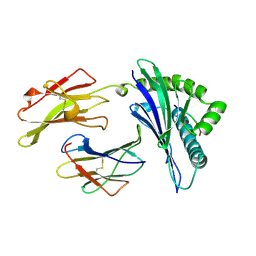 | | MHC CLASS I MOLECULE B*3501 COMPLEXED WITH PEPTIDE VPLRPMTY FROM THE NEF PROTEIN (75-82) OF HIV1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Stuart, D.I, Mcmichael, A.J, Jones, E.Y, Bell, J.I. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501.
Immunity, 4, 1996
|
|
5KLT
 
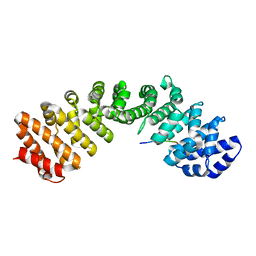 | | Prototypical P4[M]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[M]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
5KLR
 
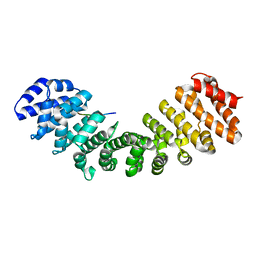 | | Prototypical P4[R]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[R]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
6X6N
 
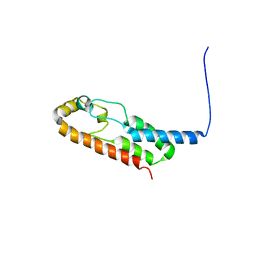 | |
7RG4
 
 | | Importin alpha2 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-1, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG5
 
 | | Importin alpha3 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-3, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
2AI8
 
 | | E.coli Polypeptide Deformylase complexed with SB-485343 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, [HYDROXY(3-PHENYLPROPYL)AMINO]METHANOL | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
5FC8
 
 | | Mouse importin alpha: Dengue 3 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nonstructural protein 5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
2AIA
 
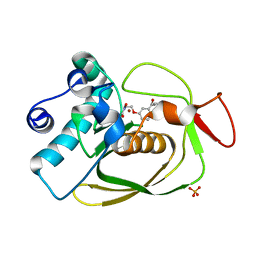 | | S.pneumoniae PDF complexed with SB-543668 | | Descriptor: | 2-(3-BENZOYLPHENOXY)ETHYL(HYDROXY)FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
2AI9
 
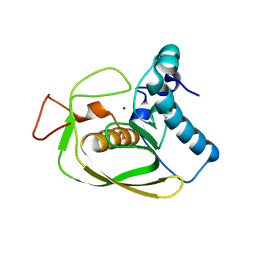 | | S.aureus Polypeptide Deformylase | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
2AIE
 
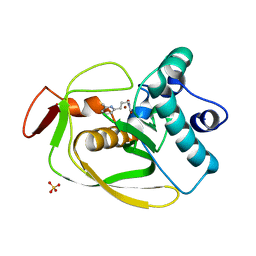 | | S.pneumoniae polypeptide deformylase complexed with SB-505684 | | Descriptor: | HYDROXY[3-(6-METHYLPYRIDIN-2-YL)PROPYL]FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
2AI7
 
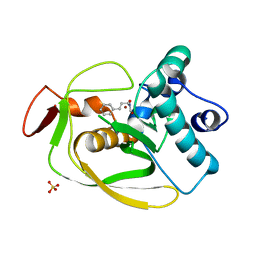 | | S.pneumoniae Polypeptide Deformylase complexed with SB-485345 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
6BVZ
 
 | | Importin alpha 3 in cargo free state | | Descriptor: | Importin subunit alpha-3 | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BVT
 
 | | Importin alpha 1 in cargo free state | | Descriptor: | Importin subunit alpha-1 | | Authors: | Smith, K.M, Tsimablyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BW0
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BVV
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6D71
 
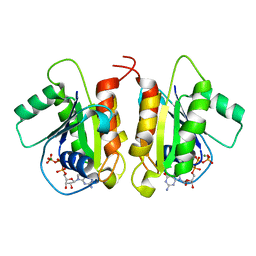 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
4G39
 
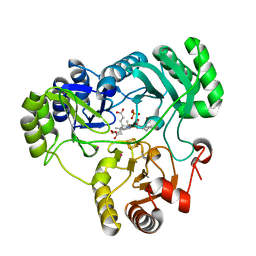 | |
4G38
 
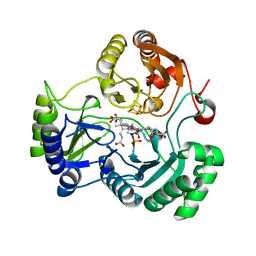 | |
3IRW
 
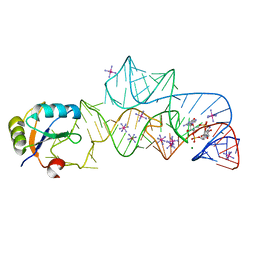 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3UD4
 
 | | The C92U mutant c-di-GMP-I riboswitch bound to GpA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*A)-3'), RNA (92-MER), ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
