3MNH
 
 | | Human Carbonic Anhydrase II Mutant K170A | | Descriptor: | Carbonic anhydrase 2, SODIUM ION, ZINC ION | | Authors: | Domsic, J.F, McKenna, R. | | Deposit date: | 2010-04-21 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and kinetic study of the extended active site for proton transfer in human carbonic anhydrase II.
Biochemistry, 49, 2010
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
 | |
8HNV
 
 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | Descriptor: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6B7K
 
 | | GH43 Endo-Arabinanase from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Endo-alpha-(1->5)-L-arabinanase | | Authors: | Farro, E.G.S, Nascimento, A.S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | GH43 endo-arabinanase from Bacillus licheniformis: Structure, activity and unexpected synergistic effect on cellulose enzymatic hydrolysis.
Int. J. Biol. Macromol., 117, 2018
|
|
4GGF
 
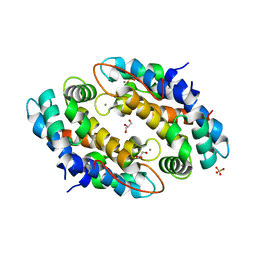 | | Crystal structure of Mn2+ bound calprotectin | | Descriptor: | CALCIUM ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Damo, S.M, Fritz, G. | | Deposit date: | 2012-08-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KS0
 
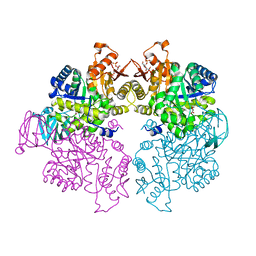 | | Pyruvate kinase (PYK) from Trypanosoma cruzi in the presence of Magnesium, oxalate and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Morgan, H.P, Zhong, W, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
3GEI
 
 | |
4JB0
 
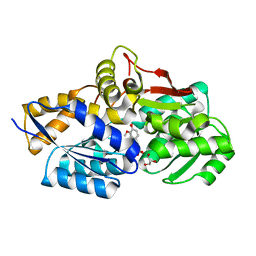 | | Rhodopseudomonas palustris (strain CGA009) Rp1789 transport protein | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, GLYCEROL, ... | | Authors: | Salmon, R. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The CouPSTU and TarPQM Transporters in Rhodopseudomonas palustris: Redundant, Promiscuous Uptake Systems for Lignin-Derived Aromatic Substrates.
Plos One, 8, 2013
|
|
4DXN
 
 | |
4EES
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
2E9Q
 
 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
4HUC
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain B and C | | Descriptor: | ACETATE ION, PROBABLE CONSERVED LIPOPROTEIN LPPS, SODIUM ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IAE
 
 | | Crystal structure of BAY 60-2770 bound to nostoc H-NOX domain | | Descriptor: | 4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid, Alr2278 protein, MALONATE ION | | Authors: | Kumar, V, van den Akker, F, Martin, F. | | Deposit date: | 2012-12-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into BAY 60-2770 Activation and S-Nitrosylation-Dependent Desensitization of Soluble Guanylyl Cyclase via Crystal Structures of Homologous Nostoc H-NOX Domain Complexes.
Biochemistry, 52, 2013
|
|
4GOB
 
 | | Low pH Crystal Structure of a reconstructed Kaede-type Red Fluorescent Protein, Least Evolved Ancestor (LEA) | | Descriptor: | Kaede-type Fluorescent Protein | | Authors: | Kim, H, Grunkemeyer, T.J, Chen, L, Fromme, R, Wachter, R.M. | | Deposit date: | 2012-08-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Acid-base catalysis and crystal structures of a least evolved ancestral GFP-like protein undergoing green-to-red photoconversion.
Biochemistry, 52, 2013
|
|
4L7T
 
 | | B. fragilis NanU | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NanU sialic acid binding protein, ... | | Authors: | Rafferty, J.B, Phansopa, C, Stafford, G.P. | | Deposit date: | 2013-06-14 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species.
Biochem.J., 458, 2014
|
|
2QCG
 
 | |
4CL2
 
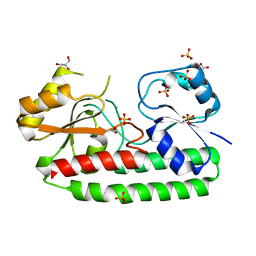 | | structure of periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Sharma, N, Selvakumar, P, Bhose, S, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of a Periplasmic Solute Binding Protein in Metal-Free, Intermediate and Metal-Bound States from Candidatus Liberibacter Asiaticus.
J.Struct.Biol., 189, 2015
|
|
5OO0
 
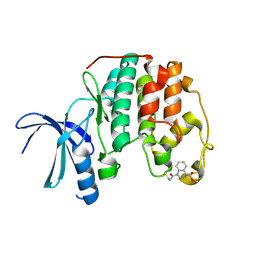 | | Cdk2(WT) covalent adduct with D28 at C177 | | Descriptor: | Cyclin-dependent kinase 2, methyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ONC
 
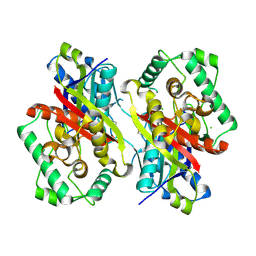 | | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis is Controlled by a Redox-Sensitive Thiol Switch | | Descriptor: | CHLORIDE ION, Steroid 3-ketoacyl-CoA thiolase | | Authors: | Schaefer, C, Kuper, J, Sampson, N.S, Kisker, C. | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch.
ACS Infect Dis, 3, 2017
|
|
5OSJ
 
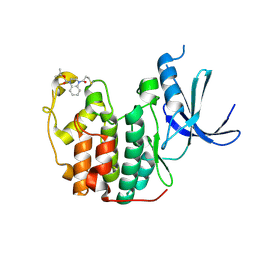 | | Cdk2(WT) with covalent adduct at C177 | | Descriptor: | Cyclin-dependent kinase 2, ~{tert}-butyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2XTT
 
 | | Bovine trypsin in complex with evolutionary enhanced Schistocerca gregaria protease inhibitor 1 (SGPI-1-P02) | | Descriptor: | ACETATE ION, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Wahlgren, W.Y, Pal, G, Kardos, J, Porrogi, P, Szenthe, B, Patthy, A, Graf, L, Katona, G. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The catalytic aspartate is protonated in the Michaelis complex formed between trypsin and an in vitro evolved substrate-like inhibitor: a refined mechanism of serine protease action.
J.Biol.Chem., 286, 2011
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5PFY
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 162) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PGD
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 177) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
