3J30
 
 | |
5ANM
 
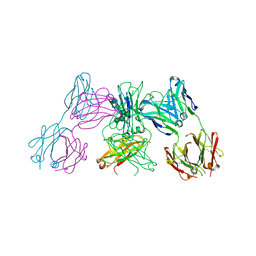 | | Crystal structure of IgE Fc in complex with a neutralizing antibody | | Descriptor: | IG EPSILON CHAIN C REGION, IMMUNOGLOBULIN G, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cohen, E.S, Dobson, C.L, Kack, H, Wang, B, Sims, D.A, Lloyd, C.O, England, E, Rees, D.G, Guo, H, Karagiannis, S.N, O'Brien, S, Persdotter, S, Ekdahl, H, Butler, R, Keyes, F, Oakley, S, Carlsson, M, Briend, E, Wilkinson, T, Anderson, I.K, Monk, P.D, vonWachenfeldt, K, Eriksson, P.O, Gould, H.J, Vaughan, T.J, May, R.D. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Ige-Neutralizing Antibody for the Treatment of Severe Uncontrolled Asthma.
Mabs, 6, 2015
|
|
3J2X
 
 | |
3J91
 
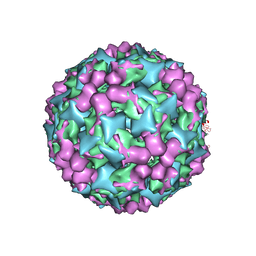 | | Cryo-electron microscopy of Enterovirus 71 (EV71) procapsid in complex with Fab fragments of neutralizing antibody 22A12 | | Descriptor: | VP0, VP1, VP3 | | Authors: | Shingler, K.L, Cifuente, J.O, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro.
J.Virol., 89, 2015
|
|
6L62
 
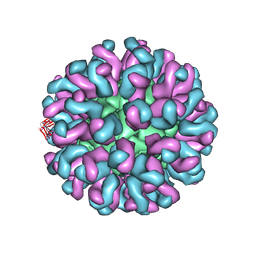 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
7MXL
 
 | |
8C7J
 
 | |
8C7V
 
 | |
5T5B
 
 | |
5T29
 
 | | Crystal structure of 10E8 Fab light chain mutant3, against the MPER region of the HIV-1 Env, in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, Antibody 10E8 FAB LIGHT CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
6XCM
 
 | |
6XCN
 
 | |
6XC2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
5SZF
 
 | | 2A10 FAB fragment 2.54 Angstoms | | Descriptor: | 2A10 antibody FAB fragment heavy chain, 2A10 antibody FAB fragment light chain, SULFATE ION | | Authors: | Jackson, C.J, Fisher, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-07-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | T-dependent B cell responses to Plasmodium induce antibodies that form a high-avidity multivalent complex with the circumsporozoite protein.
PLoS Pathog., 13, 2017
|
|
5T33
 
 | | Crystal structure of strain-specific glycan-dependent CD4 binding site-directed neutralizing antibody CAP257-RH1, in complex with HIV-1 strain RHPA gp120 core with an oligomannose N276 glycan. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CAP257-RH1 heavy chain, ... | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2092 Å) | | Cite: | Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site.
J.Virol., 90, 2016
|
|
6XC3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
7YV2
 
 | |
5ANY
 
 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
6XCA
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C105 | | Descriptor: | C105 Heavy Chain, C105 Light Chain, SULFATE ION | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.
Cell, 182, 2020
|
|
6E5P
 
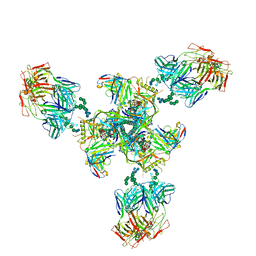 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | Descriptor: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-07-21 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
9F18
 
 | |
9F1I
 
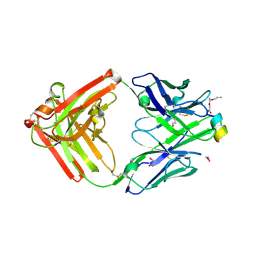 | | Crystal structure of a first-in-class antibody for alpha-1,6-fucosylated prostate-specific antigen, target bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-azanylethoxy)ethoxy]ethanoic acid, Heavy chain rabbit fab, ... | | Authors: | Halldorsson, S. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Development of a first-in-class antibody and a specific assay for alpha-1,6-fucosylated prostate-specific antigen.
Sci Rep, 14, 2024
|
|
6KYZ
 
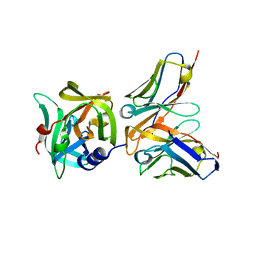 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BK2
 
 | |
