2K1M
 
 | |
5OTF
 
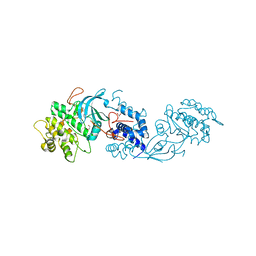 | | MRCK beta in complex with BDP-00009066 | | Descriptor: | (6~{S})-8-(3-pyrimidin-4-yl-1~{H}-pyrrolo[2,3-b]pyridin-4-yl)-1,8-diazaspiro[5.5]undecane, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent and Selective MRCK Inhibitors with Therapeutic Effect on Skin Cancer.
Cancer Res., 78, 2018
|
|
2KKQ
 
 | | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158. | | Descriptor: | Myotilin | | Authors: | Rossi, P, Shastry, R, Ciccosanti, C, Hamilton, K, Xiao, R, Acton, T.B, Swapna, G.V.T, Nair, R, Everett, J.K, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158.
To be Published
|
|
2KIN
 
 | | KINESIN (MONOMERIC) FROM RATTUS NORVEGICUS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN, SULFATE ION | | Authors: | Sack, S, Muller, J, Kozielski, F, Marx, A, Thormahlen, M, Biou, V, Mandelkow, E.-M, Brady, S.T, Mandelkow, E. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of motor and neck domains from rat brain kinesin.
Biochemistry, 36, 1997
|
|
2K3S
 
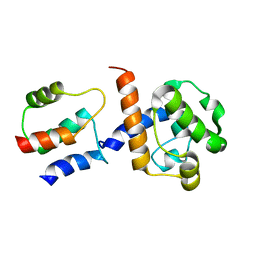 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
2KC2
 
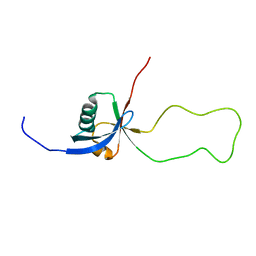 | | NMR structure of the F1 domain (residues 86-202) of the talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Elliott, P.R, Roberts, G.C.K, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2008-12-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation.
Embo J., 29, 2010
|
|
2KGB
 
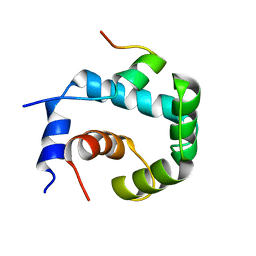 | | NMR solution of the regulatory domain cardiac F77W-Troponin C in complex with the cardiac Troponin I 144-163 switch peptide | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Mercier, P, Julien, O, Crane, M.L, Sykes, B.D. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The effect of the cosolvent trifluoroethanol on a tryptophan side chain orientation in the hydrophobic core of troponin C.
Protein Sci., 18, 2009
|
|
2KRD
 
 | | Solution Structure of the Regulatory Domain of Human Cardiac Troponin C in Complex with the Switch Region of cardiac Troponin I and W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Oleszczuk, M, Robertson, I.M, Li, M.X, Sykes, B.D. | | Deposit date: | 2009-12-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: the basis of W7 as an inhibitor of cardiac muscle contraction.
J.MOL.CELL.CARDIOL., 48, 2010
|
|
2KMA
 
 | | NMR structure of the F0F1 double domain (residues 1-202) of the talin ferm domain | | Descriptor: | Talin 1 | | Authors: | Goult, B.T, Elliott, P.R, Bate, N, Roberts, G.C, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2009-07-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation.
Embo J., 2010
|
|
4U0S
 
 | | Structure of Eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Katan, M, Bunney, T.D. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4KCA
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4FFB
 
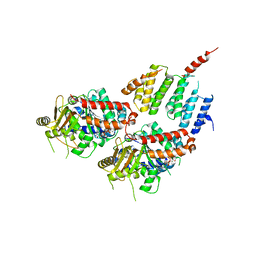 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
8OVT
 
 | |
5L3T
 
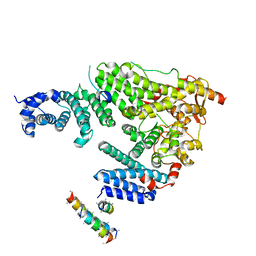 | | Structure of the Saccharomyces cerevisiae TREX-2 complex | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Stewart, M, Aibara, S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.927 Å) | | Cite: | The Sac3 TPR-like region in the Saccharomyces cerevisiae TREX-2 complex is more extensive but independent of the CID region.
J. Struct. Biol., 195, 2016
|
|
2D88
 
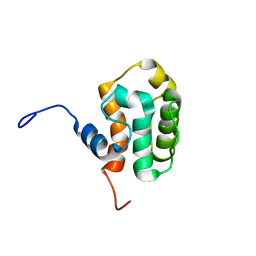 | | Solution structure of the CH domain from human MICAL-3 protein | | Descriptor: | Protein MICAL-3 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from human MICAL-3 protein
To be Published
|
|
7TQ7
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
3LC4
 
 | |
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ8
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
3L8Q
 
 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
5M6C
 
 | | CRYSTAL STRUCTURE OF T71N MUTANT OF HUMAN HIPPOCALCIN | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Helassa, N, Antonyuk, S.V, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
8QVU
 
 | | Crystal Structure of ligand ACBI3 in complex with KRAS G12D C118S GDP and pVHL:ElonginC:ElonginB complex | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Wijaya, A.J, Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
