5M6C
 
 | | CRYSTAL STRUCTURE OF T71N MUTANT OF HUMAN HIPPOCALCIN | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Helassa, N, Antonyuk, S.V, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
7ZRP
 
 | | 2.65 Angstrom crystal structure of Ca/CaM:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
7ZRQ
 
 | | 1.68 Angstrom crystal structure of Ca/CaM-E140G:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
5G4P
 
 | | Crystal structure of human hippocalcin at 2.4 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
5G58
 
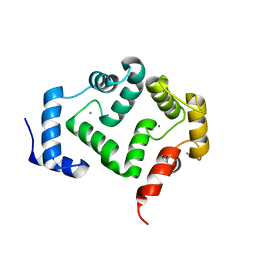 | | Crystal structure of A190T mutant of human hippocalcin AT 2.5 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-22 | | Release date: | 2017-05-03 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
6XY3
 
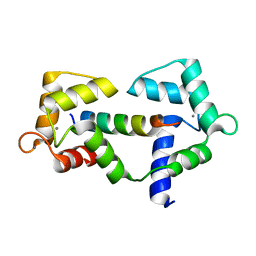 | |
6XXX
 
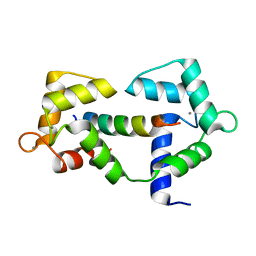 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
6XXF
 
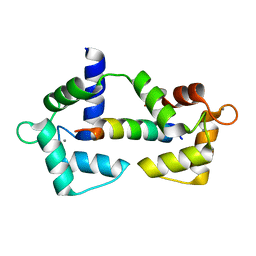 | |
