4U3J
 
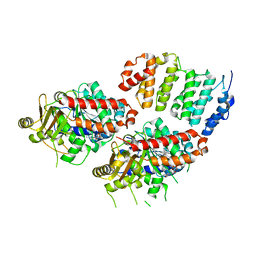 | | TOG2:alpha/beta-tubulin complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Rice, L.M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Elife, 3, 2014
|
|
4FFB
 
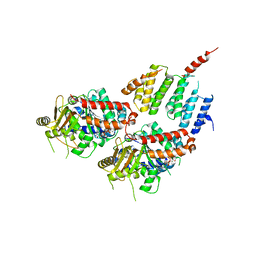 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
5IEX
 
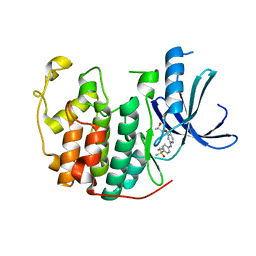 | | Crystal structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-hydroxy-1-methylpropyl]oxy}- pyrimidin-2-yl)amino]phenyl}-S-cyclopropylsulfoximide bound to CDK2 | | Descriptor: | (2R,3R)-3-[(5-bromo-2-{[4-(S-cyclopropylsulfonimidoyl)phenyl]amino}pyrimidin-4-yl)oxy]butan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IF1
 
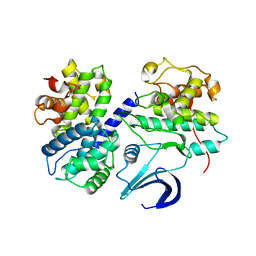 | | Crystal structure apo CDK2/cyclin A | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IEV
 
 | | Crystal structure of BAY 1000394 (Roniciclib) bound to CDK2 | | Descriptor: | Cyclin-dependent kinase 2, Roniciclib | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IEY
 
 | | Crystal structure of a CDK inhibitor bound to CDK2 | | Descriptor: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
