2Q5D
 
 | |
2OT8
 
 | |
2QMR
 
 | | Karyopherin beta2/transportin | | Descriptor: | Transportin-1 | | Authors: | Cansizoglu, A.E, Chook, Y.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational heterogeneity of karyopherin beta2 is segmental
Structure, 15, 2007
|
|
1O6P
 
 | |
1O6O
 
 | |
1QGR
 
 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
8DYO
 
 | | Cryo-EM structure of Importin-4 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | Authors: | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8F19
 
 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-5, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F7A
 
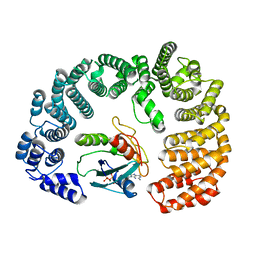 | | Cryo-EM structure of Importin-9 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-9, ... | | Authors: | Bernardes, N.E, Chook, Y.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1E
 
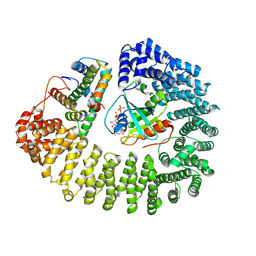 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) and H2A-H2B | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Histone H2A.2, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0X
 
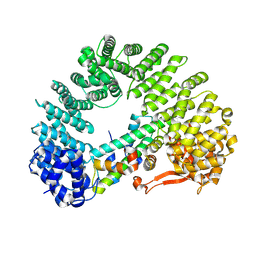 | | Cryo-EM structure of Kap114 bound to H2A-H2B | | Descriptor: | Histone H2A.2, Histone H2B.2, Importin subunit beta-5 | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1QGK
 
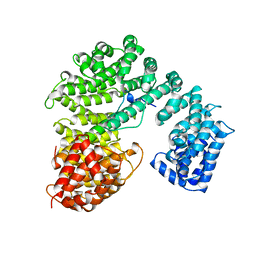 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QBK
 
 | |
6M6X
 
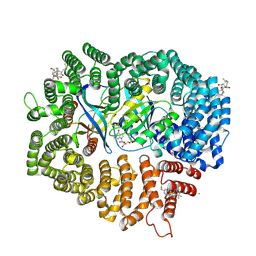 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6LQ9
 
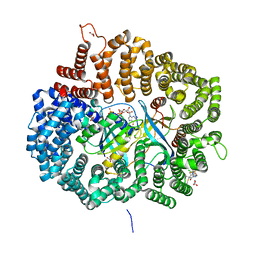 | | S109 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (3~{R},4~{S})-1-[[6-chloranyl-5-(trifluoromethyl)pyridin-2-yl]amino]-3,4-dimethyl-pyrrolidine-2,5-dione, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
6M60
 
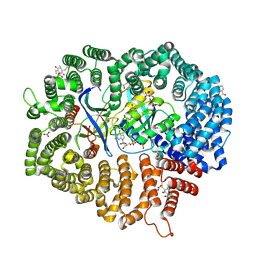 | | Plumbagin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
3A6P
 
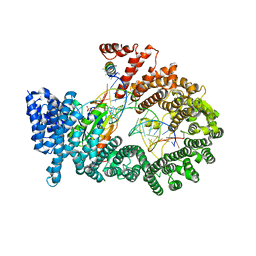 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-08 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
6N88
 
 | |
3M1I
 
 | |
3NC1
 
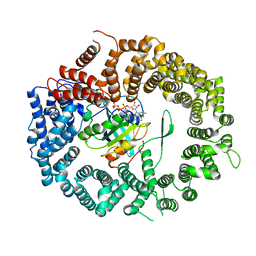 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2Z5M
 
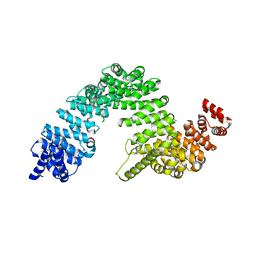 | | Complex of Transportin 1 with TAP NLS, crystal form 2 | | Descriptor: | Nuclear RNA export factor 1, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
5DHF
 
 | |
5DIS
 
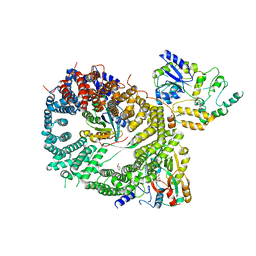 | | Crystal structure of a CRM1-RanGTP-SPN1 export complex bound to a 113 amino acid FG-repeat containing fragment of Nup214 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Monecke, T, Port, S.A, Dickmanns, A, Kehlenbach, R.H, Ficner, R. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export.
Cell Rep, 13, 2015
|
|
5DH9
 
 | |
6N89
 
 | |
