9FZH
 
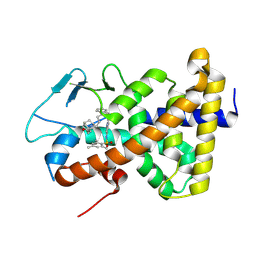 | | Crystal structure of the hPXR-LBD in complex with compound JMV6995 | | Descriptor: | 2-(1-adamantyl)-~{N}-[7-[1-(phenylmethyl)-5-[(2,4,6-trimethylphenyl)sulfonylamino]benzimidazol-2-yl]heptyl]ethanamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Bourguet, W. | | Deposit date: | 2024-07-05 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A two-in-one expression construct for biophysical and structural studies of the human pregnane X receptor ligand-binding domain, a pharmaceutical and environmental target.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
3HXV
 
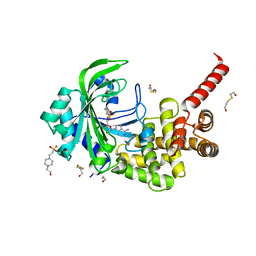 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
8J8J
 
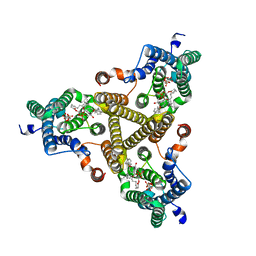 | | Membrane bound PRTase, C3 symmetry, donor bound | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Decaprenyl-phosphate phosphoribosyltransferase, ... | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
7R14
 
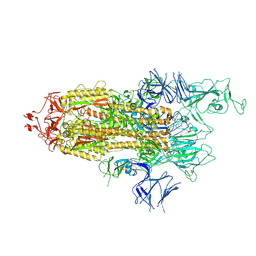 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
1AKS
 
 | | CRYSTAL STRUCTURE OF THE FIRST ACTIVE AUTOLYSATE FORM OF THE PORCINE ALPHA TRYPSIN | | Descriptor: | ALPHA TRYPSIN, CALCIUM ION | | Authors: | Johnson, A, Krishnaswamy, S, Sundaram, P.V, Pattabhi, V. | | Deposit date: | 1996-07-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure at 1.8 A resolution of an active autolysate form of porcine alpha-trysoin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4ZK8
 
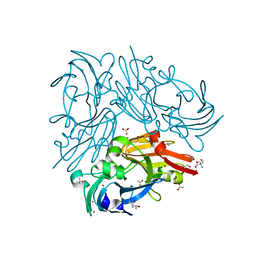 | | Copper-containing nitrite reductase from thermophilic bacterium Geobacillus thermodenitrificans (Re-refined) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fukuda, Y, Inoue, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the function of a thermostable copper-containing nitrite reductase
J.Biochem., 155, 2014
|
|
8W96
 
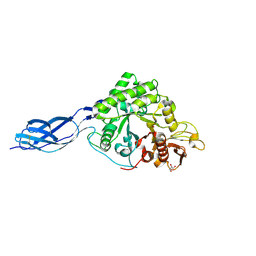 | | SmChiA with diacetyl chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Tanaka, Y, Nakamura, A. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Product inhibition slow down the moving velocity of processive chitinase and sliding-intermediate state blocks re-binding of product.
Arch.Biochem.Biophys., 752, 2024
|
|
5MZO
 
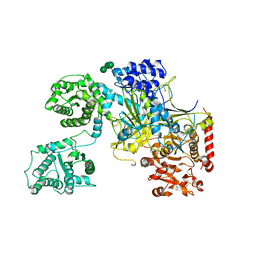 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (open conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4A5N
 
 | | Redoxregulator HypR in its reduced form | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, UNCHARACTERIZED HTH-TYPE TRANSCRIPTIONAL REGULATOR YYBR | | Authors: | Palm, G.J, Waack, P, Read, R.J, Hinrichs, W. | | Deposit date: | 2011-10-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights Into the Redox-Switch Mechanism of the Marr/Duf24-Type Regulator Hypr
Nucleic Acids Res., 40, 2012
|
|
5GTN
 
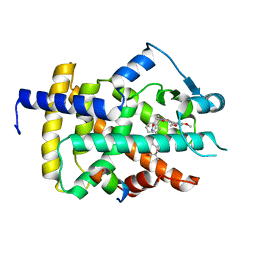 | | Human PPARgamma ligand binding dmain complexed with R35 | | Descriptor: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
2VXV
 
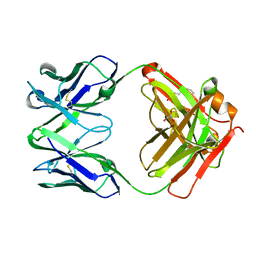 | | Crystal structure of human IgG ABT-325 Fab Fragment | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, HUMAN IGG ABT-325 | | Authors: | Argiriadi, M.A, Xiang, T, Wu, C, Ghayur, T, Borhani, D.W. | | Deposit date: | 2008-07-10 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Unusual Water-Mediated Antigenic Recognition of the Proinflammatory Cytokine Interleukin-18.
J.Biol.Chem., 284, 2009
|
|
7Y3H
 
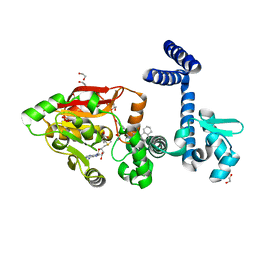 | | Crystal Structure of Diels-Alderase ApiI in complex with SAM and product | | Descriptor: | 1,2-ETHANEDIOL, 3-[[(1R,2R,4aS,8aR)-2-methyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]carbonyl]-5-(4-hydroxyphenyl)-4-oxidanyl-1H-pyridin-2-one, ApiI, ... | | Authors: | Zhou, J.H, Lu, J.Y. | | Deposit date: | 2022-06-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Diels-Alderase ApiI in complex with SAM and product
To Be Published
|
|
8J9H
 
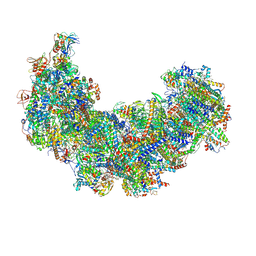 | | Cryo-EM structure of Euglena gracilis respiratory complex I, deactive state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
5DJM
 
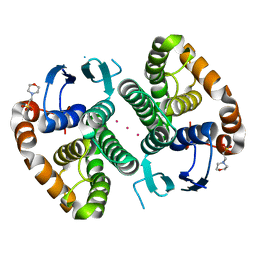 | |
4RIU
 
 | |
2Y7B
 
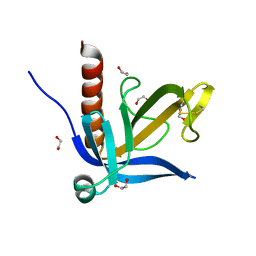 | | Crystal structure of the PH domain of human Actin-binding protein anillin ANLN | | Descriptor: | 1,2-ETHANEDIOL, ACTIN-BINDING PROTEIN ANILLIN | | Authors: | Vollmar, M, Wang, J, Krojer, T, Elkins, J, Filippakopoulos, P, Ugochukwu, E, Cocking, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Ph Domain of Human Actin-Binding Protein Anillin Anln
To be Published
|
|
7K8X
 
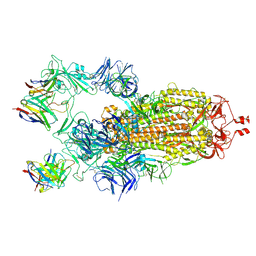 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
2VO4
 
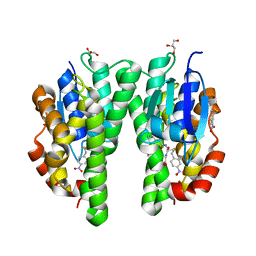 | | Glutathione transferase from Glycine max | | Descriptor: | 2,4-D INDUCIBLE GLUTATHIONE S-TRANSFERASE, 4-NITROPHENYL METHANETHIOL, GLYCEROL, ... | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and Functional Characterization of the Fluorodifen-Inducible Glutathione Transferase from Glycine Max Reveals an Active Site Topography Suited for Diphenylether Herbicides and a Novel L-Site.
J.Mol.Biol., 385, 2009
|
|
1GUV
 
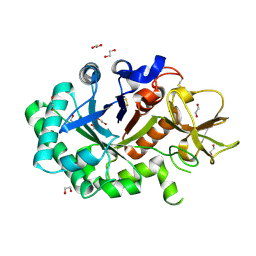 | | Structure of human chitotriosidase | | Descriptor: | 1,2-ETHANEDIOL, CHITOTRIOSIDASE | | Authors: | Von Moeller, H, Houston, D, Boot, R.G, Aerts, J.M.F.G, Van Aalten, D.M.F. | | Deposit date: | 2002-01-31 | | Release date: | 2003-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Human Chitotriosidase - Implications for Specific Inhibitor Design and Function of Mammalian Chitinase-Like Lectins
J.Biol.Chem., 277, 2002
|
|
5JMS
 
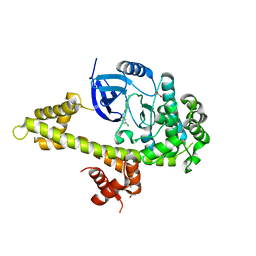 | | Crystal structure of TgCDPK1 bound to CGP060476 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-domain protein kinase 1, ... | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TgCDPK1bount to CGP060476
To Be Published
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
