6C1S
 
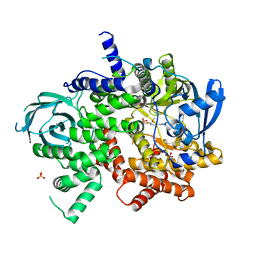 | | Phosphoinositide 3-Kinase gamma bound to an pyrrolopyridinone Inhibitor | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, {4-[2-(5,6-dimethoxypyridin-3-yl)-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-1H-pyrazol-1-yl}acetonitrile | | Authors: | Jacobs, M.D, Griffin, J.P. | | Deposit date: | 2018-01-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Synthesis of a Novel Series of Orally Bioavailable, CNS-Penetrant, Isoform Selective Phosphoinositide 3-Kinase gamma (PI3K gamma ) Inhibitors with Potential for the Treatment of Multiple Sclerosis (MS).
J. Med. Chem., 61, 2018
|
|
1KEM
 
 | |
7VVA
 
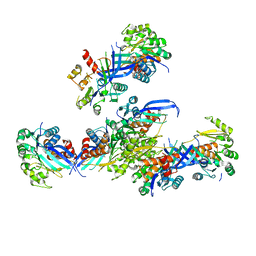 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) from Escherichia coli strain B | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75029182 Å) | | Cite: | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7M4L
 
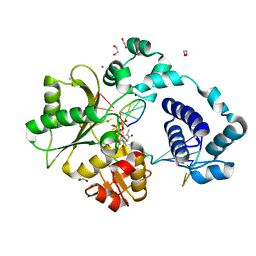 | | DNA Polymerase Lambda, TTPaS:At Mn2+ Product State Ternary Complex, 60 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*(YQS))-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7XO7
 
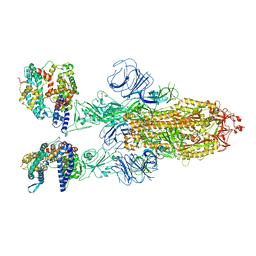 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
5UIX
 
 | |
6G2H
 
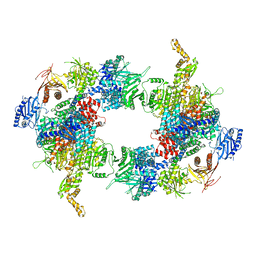 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
4U5C
 
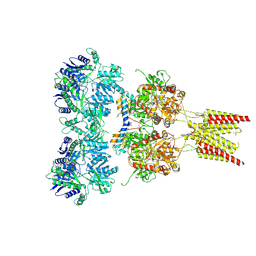 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist FW and postitive modulator (R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6883 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
5B6S
 
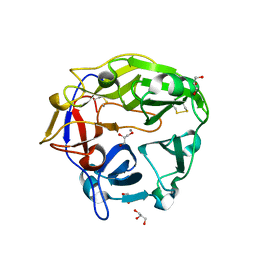 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
6S25
 
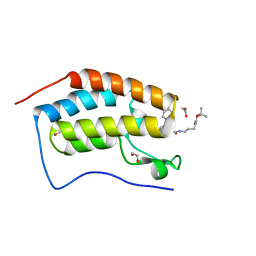 | | Crystal Structure of the first bromodomain of BRD4 in complex with a benzodiazepine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{tert}-butyl ~{N}-[3-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]propyl]carbamate | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with a benzodiazepine ligand
To Be Published
|
|
9NGE
 
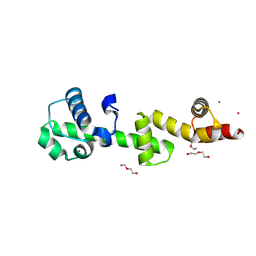 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 rigidly fused to a double trigger variant of the 1TEL crystallization chaperone | | Descriptor: | DI(HYDROXYETHYL)ETHER, POTASSIUM ION, SODIUM ION, ... | | Authors: | Averett, J.C, Bradford, M.J, Averett, B.J, Hansen, D, Doukov, T, Moody, J.D. | | Deposit date: | 2025-02-22 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The ubiquitin-associated domain of human thirty-eight negative kinase-1 rigidly fused to a double trigger variant of the 1TEL crystallization chaperone
To Be Published
|
|
9M1F
 
 | | Crystal structure of E. coli tryptophanyl-tRNA synthetase complexed with chuangxinmycin and ATP in closed-closed state | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Ren, Y, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2025-02-25 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
4U69
 
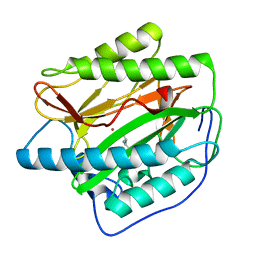 | | HsMetAP complex with (1-amino-2-methylpentyl)phosphonic acid | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 1, POTASSIUM ION, ... | | Authors: | Arya, T, Addlagatta, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the Molecular Basis of Inhibitor Selectivity between the Human and Streptococcal Type I Methionine Aminopeptidases
J.Med.Chem., 58, 2015
|
|
7M6Q
 
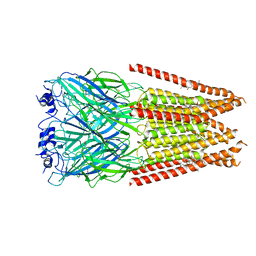 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
5IR2
 
 | | Crystal structure of novel cellulases from microbes associated with the gut ecosystem | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cellulase, ... | | Authors: | Chang, C, Mack, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of novel cellulases from microbes associated with the gut ecosystem
To Be Published
|
|
4UC1
 
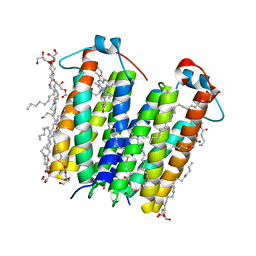 | | High resolution crystal structure of translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, METHOXY-ETHOXYL, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
6S6Y
 
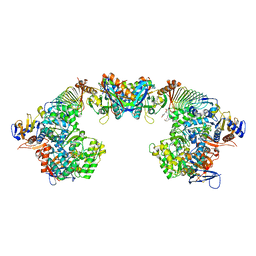 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5AF7
 
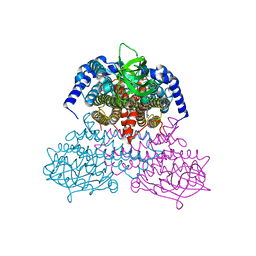 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U70
 
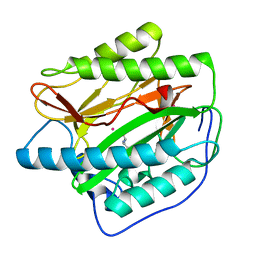 | |
3Q5Y
 
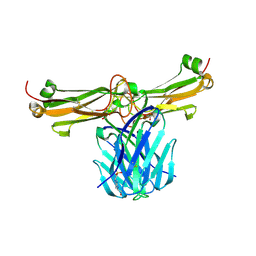 | | V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Q, Zhang, H, Wang, J.-H. | | Deposit date: | 2010-12-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved hydrophobic patch on Vbeta domains revealed by TCRbeta chain crystal structures: implications for pre-TCR dimerization
Front Immunol, 2, 2011
|
|
5ZFL
 
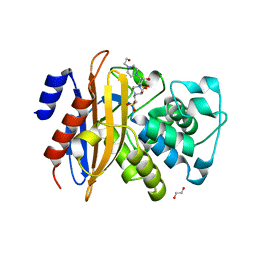 | | Crystal structure of beta-lactamase PenP mutant E166Y | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydrolytic water molecule of Class A beta-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis.
Sci Rep, 10, 2020
|
|
5A6O
 
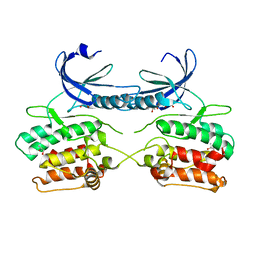 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6EI4
 
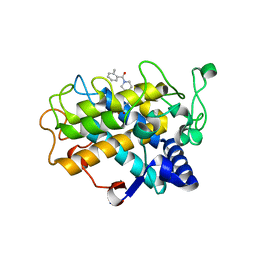 | | Crystal Structure of tyrosinase from Bacillus megaterium with B5N inhibitor in the active site | | Descriptor: | COPPER (II) ION, Tyrosinase, [4-[(4-fluorophenyl)methyl]piperazin-1-yl]-(2-methylphenyl)methanone | | Authors: | Deri, B, Gitto, R, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-09-17 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Tyrosinase: Development and Structural Insights of Novel Inhibitors Bearing Arylpiperidine and Arylpiperazine Fragments.
J. Med. Chem., 61, 2018
|
|
5XPI
 
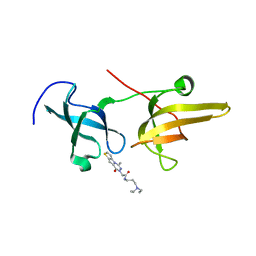 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
5I2K
 
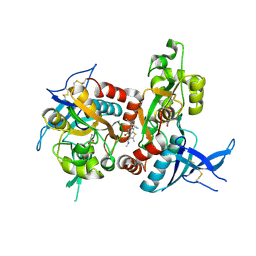 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
