6FFH
 
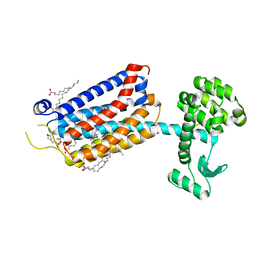 | | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(3-chlorophenyl)-3-(3-methyl-5-oxidanylidene-4~{H}-imidazol-2-yl)urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
7MIU
 
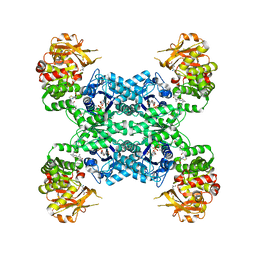 | | Mouse CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIH
 
 | | Human CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SMH
 
 | |
8EW3
 
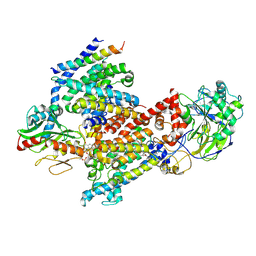 | | Cryo EM structure of Vibrio cholerae NQR | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit A, ... | | Authors: | Fuller, J.R, Juarez, O. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.65159 Å) | | Cite: | Novel cofactor binding motifs, electron transfer and ion pumping mechanisms of the respiratory complex NQR
To Be Published
|
|
8SP1
 
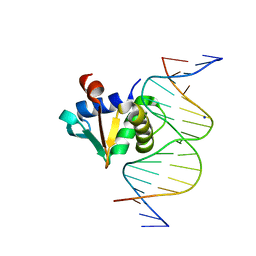 | |
4RX5
 
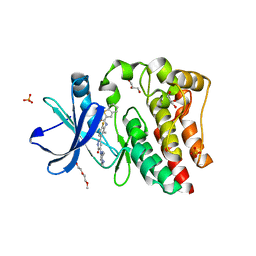 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 23 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, N-(6-fluoro-2-methyl-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl)-1-benzothiophene-2-carboxamide, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | Discovery of highly potent and selective Bruton's tyrosine kinase inhibitors: Pyridazinone analogs with improved metabolic stability.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HYA
 
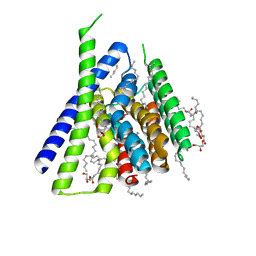 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchangerNCX_Mj soaked with 150 mM Na+ and nominal Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, CALCIUM ION, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JHA
 
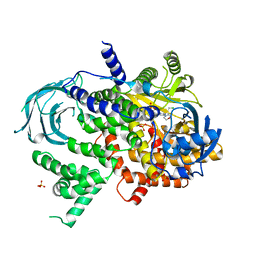 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin2 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)pyrimidin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
4TU8
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5A6 DNA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DA*U)-3'), ... | | Authors: | MCLAUGHLIN, K.J, JENKINS, J.L, Agrawal, A.A, KIELKOPF, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5A57
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT | | Descriptor: | (Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
9L6A
 
 | | Crystal structure of KRas G12D (GDP) in complex with compound 1 | | Descriptor: | 6-cyclopropyl-4-[(1~{S},4~{S})-2,5-diazabicyclo[2.2.1]heptan-2-yl]-7-(6-fluoranyl-5-methyl-1~{H}-indazol-4-yl)-2-(oxan-4-yloxy)-8-phenylmethoxy-quinazoline, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2024-12-24 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Discovery of KRAS(G12D) selective degrader ASP3082.
Commun Chem, 8, 2025
|
|
3IB8
 
 | | Crystal structure of full length Rv0805 in complex with 5'-AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability
J.Biol.Chem., 284, 2009
|
|
8SGE
 
 | | KLHDC2 Kelch Domain with ligand KDRLKZ-1 | | Descriptor: | GLYCEROL, Kelch domain-containing protein 2, [(5P)-5-{3-[(2R)-butan-2-yl]-7-[(2-methoxyethoxy)carbonyl]-2-oxo-5,6,7,8-tetrahydro-1,7-naphthyridin-1(2H)-yl}-2-oxopyridin-1(2H)-yl]acetic acid | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3NW3
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
7ZVX
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
5HNK
 
 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8EVU
 
 | | Cryo EM structure of Vibrio cholerae NQR | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit A, ... | | Authors: | Fuller, J.R, Juarez, O. | | Deposit date: | 2022-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5804 Å) | | Cite: | Novel cofactor binding motifs, electron transfer and ion pumping mechanisms of the respiratory complex NQR
To Be Published
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
6UOL
 
 | |
6R27
 
 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
8AQU
 
 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8V9Q
 
 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
5AE1
 
 | | Ether Lipid-Generating Enzyme AGPS in complex with inhibitor ZINC69435460 | | Descriptor: | (3-(2-FLUOROPHENYL)-N-(1-(2-OXO-2,3-DIHYDRO-1H-BENZO[D]IMIDAZOL-5-YL)ETHYL)BUTANAMIDE), ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
6IPW
 
 | | Crystal structure of CqsB2 from Streptomyces exfoliatus in complex with the product, 1-(2-hydroxypropyl)-2-methyl-carbazole-3,4-dione | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB2 | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2018-11-05 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Unprecedented Cyclization Mechanism in the Biosynthesis of Carbazole Alkaloids in Streptomyces.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
