3GKZ
 
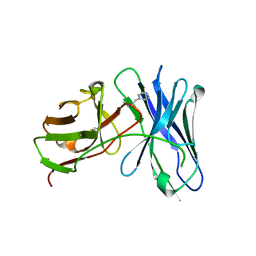 | | Crystal structures of a therapeutic single chain antibody in complex methamphetamine | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, anti-methamphetamine single chain Fv | | Authors: | Celikel, R, Peterson, E.C, Owens, M, Varughese, K.I. | | Deposit date: | 2009-03-11 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a therapeutic single chain antibody in complex with two drugs of abuse-Methamphetamine and 3,4-methylenedioxymethamphetamine.
Protein Sci., 18, 2009
|
|
1LTR
 
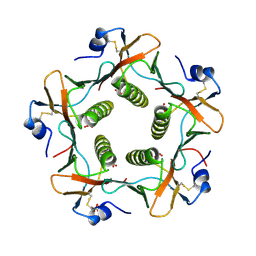 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HUMAN HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SULFATE ION | | Authors: | Matkovic-Calogovic, D, Loreggian, A, Palu, G, Zanotti, G. | | Deposit date: | 1998-07-31 | | Release date: | 1999-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
4U19
 
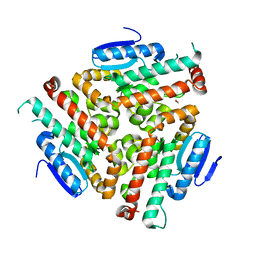 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase V349A mutant (ISOA-ECI2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
8A8O
 
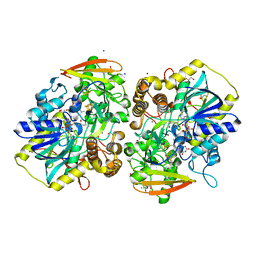 | | PAPS reductase from Methanothermococcus thermolithotrophicus refined to 1.45 A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Alpha-subunit of the PAPS reductase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Jespersen, M, Wagner, T. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Assimilatory sulfate reduction in the marine methanogen Methanothermococcus thermolithotrophicus.
Nat Microbiol, 8, 2023
|
|
7MLO
 
 | | Crystal structure of ricin A chain in complex with 5-mesitylthiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,4,6-trimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
8TCA
 
 | | Structure of a mouse IgG antibody antigen-binding fragment (Fab) targeting N6-methyladenosine (m6A), an RNA modification, no ligand | | Descriptor: | 1,2-ETHANEDIOL, m6A binding IgG Fab, heavy chain, ... | | Authors: | Angelo, M.C, Aoki, S.T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | In silico lambda-dynamics predicts protein binding specificities to modified RNAs.
Nucleic Acids Res., 53, 2025
|
|
7ZLS
 
 | | co-crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
5U2P
 
 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
8Y64
 
 | | Crystal structure of open state ferulic acid decarboxylase from Saccharomyces cerevisiae, F397V/I398L/T438P/P441V mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ferulic acid decarboxylase 1, GLYCEROL | | Authors: | Feng, Y.B, Song, X, Zhu, X.N. | | Deposit date: | 2024-02-01 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integrated strategies for engineering ferulic acid decarboxylase with tunnel conformation and substrate pocket for adapting non-natural substrates.
Biochem Eng J, 213, 2025
|
|
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
8A7K
 
 | | PcIDS1 in complex with Mg2+/Mn2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6YB8
 
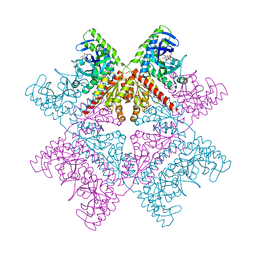 | | Human octameric PAICS in complex with CAIR and SAICAR | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
6QOK
 
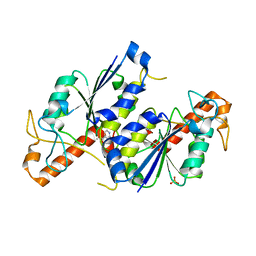 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 14 (Methyl 2-aminopyridine-4-carboxylate) | | Descriptor: | SULFATE ION, methyl 2-azanylpyridine-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
8HQ3
 
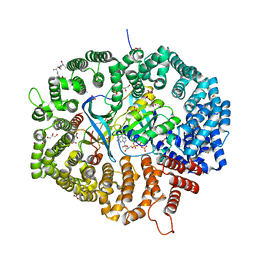 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
5NG9
 
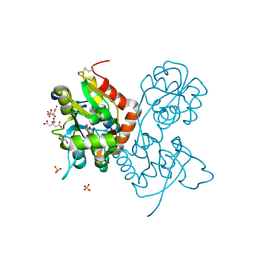 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist CIP-AS at 1.15 A resolution. | | Descriptor: | (2~{S},3~{R},4~{R})-3-(carboxycarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylic acid, (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CITRATE ANION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Winther, S, Kastrup, J.S. | | Deposit date: | 2017-03-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
9I76
 
 | |
9DUE
 
 | | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-07-081a | | Descriptor: | 3-chloro-6-(4-methylpiperazin-1-yl)-4-(pyridin-4-yl)pyridazine, BORIC ACID, Death-associated protein kinase 1, ... | | Authors: | Minasov, G, Winsor, J, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-07-081a
To Be Published
|
|
5NH0
 
 | |
6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7XFA
 
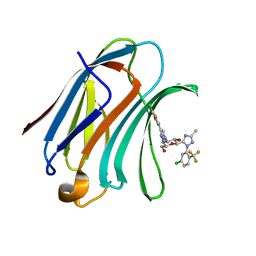 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Shukla, J, Raman, S, Ghosh, K. | | Deposit date: | 2022-04-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
7BGU
 
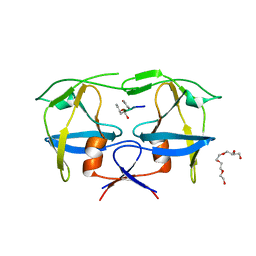 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
4RYY
 
 | |
5NRH
 
 | |
