8WS3
 
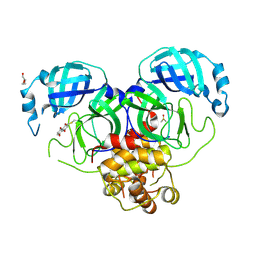 | | Crystal structure of SARS-CoV-2 Main Protease (Mpro) with covalent inhibitor 5,8-Dihydroxy-1,4-naphthoquinone | | Descriptor: | 3C-like proteinase nsp5, 5,8-bis(oxidanyl)naphthalene-1,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Wu, D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors of SARS-CoV-2 main protease
To Be Published
|
|
8E1A
 
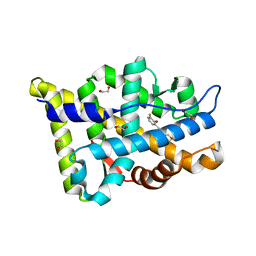 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
4RN7
 
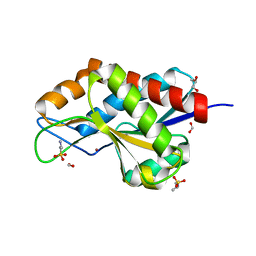 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
8Z53
 
 | | Arabidopsis Dwarf14 (AtD14) co-crystallized in the presence of Zaxinone | | Descriptor: | (7E)-6-methyl-8-[(4S)-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octa-3,5,7-trien-2-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arold, S.T, Shahul Hameed, U.F. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arabidopsis response to the apocarotenoid zaxinone involves interference with strigolactone signaling via binding to DWARF14.
Nat Commun, 16, 2025
|
|
8FV2
 
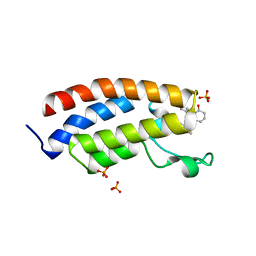 | | Bromodomain of CBP liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
7USJ
 
 | | BRD4-BD2 in complex with SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
4TK5
 
 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7UI0
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UHZ
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
6WNK
 
 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
8SU2
 
 | |
8FUP
 
 | | Bromodomain of CBP liganded with BMS-536924 and CCS-1477 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
8FVF
 
 | | Bromodomain of EP300 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Histone acetyltransferase p300, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXE
 
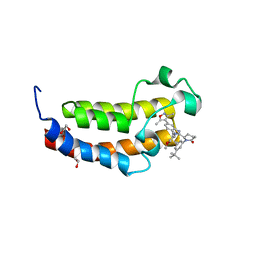 | | Bromodomain of CBP liganded with iCBP6 | | Descriptor: | (6S)-1-(3-tert-butylphenyl)-6-{(5P)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
6R8C
 
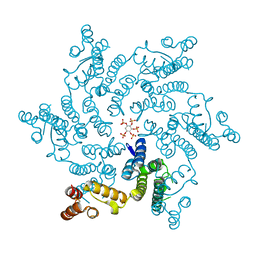 | | HIV capsid hexamer with IP5 ligand | | Descriptor: | Gag polyprotein, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2019-04-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | HIV hexamer
to be published
|
|
6ZBB
 
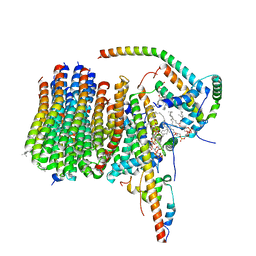 | | bovine ATP synthase Fo domain | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5Y8X
 
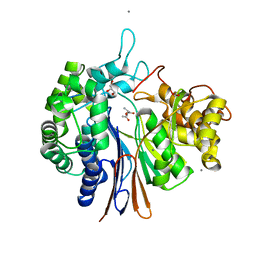 | | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumari, S, Goel, M, Pal, R, Gupta, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine
To Be Published
|
|
8FXO
 
 | | Bromodomain of CBP liganded with iCBP8 | | Descriptor: | (6S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
9GIU
 
 | | Cryo-EM structure of human SLC45A4 in detergent | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Solute carrier family 45 member 4 | | Authors: | Markusson, S, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-08-19 | | Release date: | 2025-07-30 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SLC45A4 is a pain gene encoding a neuronal polyamine transporter.
Nature, 646, 2025
|
|
8VX3
 
 | | Structure of an exported small alarmone synthetase from Streptomyces albidoflavus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Ahmad, S, Tsang, K.K, Schiefer, V, Kim, Y, Whitney, J.C. | | Deposit date: | 2024-02-03 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a (p)ppApp synthetase from Streptomyces albidoflavus
To Be Published
|
|
8UIW
 
 | | yjdF riboswitch from R. gauvreauii in complex with chelerythrine bound to Fab BL3-6 S97N | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7P19
 
 | |
6X10
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with haloperidol | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-17 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
7OWB
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
