3DOP
 
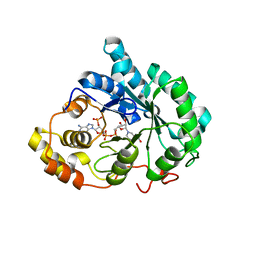 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-dihydrotestosterone, Resolution 2.00A | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 5-beta-DIHYDROTESTOSTERONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-07-05 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and catalytic mechanism of human steroid 5beta-reductase (AKR1D1)
Mol.Cell.Endocrinol., 301, 2009
|
|
3FJN
 
 | |
3EAU
 
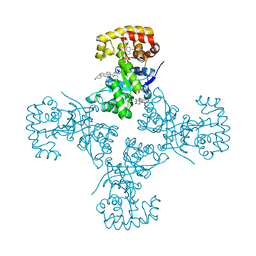 | | Voltage-dependent K+ channel beta subunit in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
3G5E
 
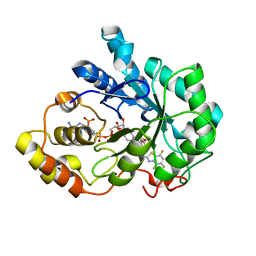 | | Human aldose reductase complexed with IDD 740 inhibitor | | Descriptor: | 2-(3-((4,5,7-trifluorobenzo[d]thiazol-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridin-1-yl)acetic acid, Aldose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Podjarny, A.D, Van Zandt, M.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of [3-(4,5,7-trifluoro-benzothiazol-2-ylmethyl)-pyrrolo[2,3-b]pyridin-1-yl]acetic acids as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GHU
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Forth stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3FX4
 
 | | Porcine aldehyde reductase in ternary complex with inhibitor | | Descriptor: | Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with a 5-arylidene-2,4-thiazolidinedione aldose reductase inhibitor.
Eur.J.Med.Chem., 45, 2010
|
|
3GHS
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Second stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3G1R
 
 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | Descriptor: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
3EB4
 
 | | Voltage-dependent K+ channel beta subunit (I211R) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
3GHT
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Third stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHR
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. First stage of radiation damage | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3CV7
 
 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
3GUG
 
 | |
3H7U
 
 | | Crystal structure of the plant stress-response enzyme AKR4C9 | | Descriptor: | ACETATE ION, Aldo-keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | White, S.A, Simpson, P.J, Ride, J.P. | | Deposit date: | 2009-04-28 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress.
J.Mol.Biol., 392, 2009
|
|
3H4G
 
 | | Structure of aldehyde reductase holoenzyme in complex with potent aldose reductase inhibitor Fidarestat: Implications for inhibitor binding and selectivity | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-04-20 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of aldehyde reductase holoenzyme in complex with the potent aldose reductase inhibitor fidarestat: implications for inhibitor binding and selectivity
J.Med.Chem., 48, 2005
|
|
3LN3
 
 | |
3LBO
 
 | | Human aldose reductase mutant T113C complexed with IDD594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-08 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LEN
 
 | | Human Aldose Reductase mutant T113S complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-15 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3LQL
 
 | | Human Aldose Reductase mutant T113A complexed with IDD 594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-02-09 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LD5
 
 | | Human aldose reductase mutant T113S complexed with IDD594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LZ3
 
 | | Human aldose reductase mutant T113S complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
2IPJ
 
 | | Crystal structure of h3alpha-hydroxysteroid dehydrogenase type 3 mutant Y24A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
2IPW
 
 | | Crystal Structure of C298A W219Y Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IQD
 
 | | Crystal Structure of Aldose Reductase complexed with Lipoic Acid | | Descriptor: | Aldose reductase, LIPOIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Carlson, E, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
3LZ5
 
 | | Human aldose reductase mutant T113V complexed with IDD594 | | Descriptor: | Aldose Reductase, CITRIC ACID, IDD594, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
