8DW1
 
 | | Crystal structure of a host-guest complex with 5'-CTTAGTTATAACTAAG-3' | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*CP*TP*AP*AP*G)-3'), reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-07-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
8DVX
 
 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
7YNE
 
 | |
8DVN
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVL
 
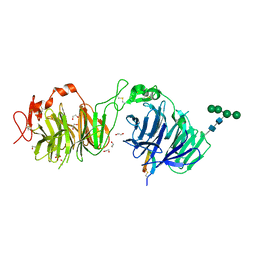 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.18 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVM
 
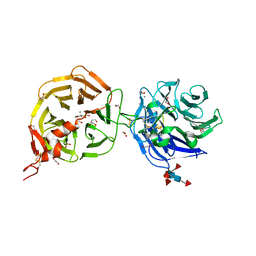 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
7YMS
 
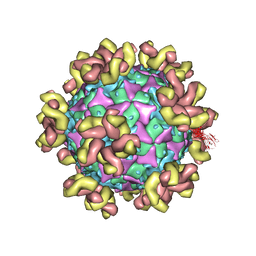 | |
8AK3
 
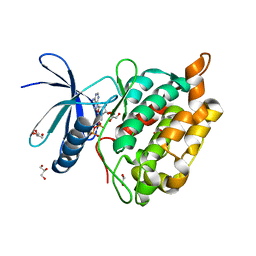 | |
8AK2
 
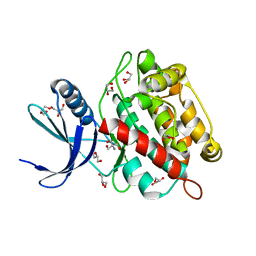 | | Drosophila melanogaster UNC89 Protein Kinase Domain 1 (apo) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Obscurin | | Authors: | Dorendorf, T, Zacharchenko, T, Mayans, O. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PK1 from Drosophila obscurin is an inactive pseudokinase with scaffolding properties.
Open Biology, 13, 2023
|
|
7YMJ
 
 | | Cryo-EM structure of alpha1AAR-Nb6 complex bound to tamsulosin | | Descriptor: | Nb6, Tamsulosin, alpha1A-adrenergic receptor | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YMH
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | | Descriptor: | Nb29, Noradrenaline, alpha1A-adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YLW
 
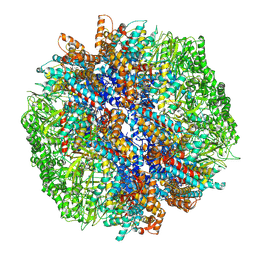 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YM8
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to oxymetazoline | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Oxymetazoline, alpha1A adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
8AJ4
 
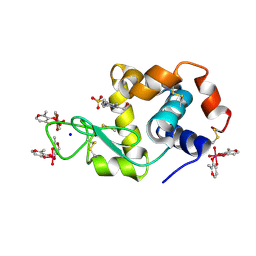 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A') | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], Lysozyme, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ3
 
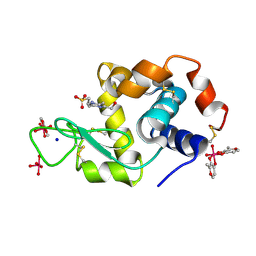 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8,8,8-tetrakis($l^{1}-oxidanyl)-2-methyl-3,7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1,5-diene, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ5
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure B) | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8DTN
 
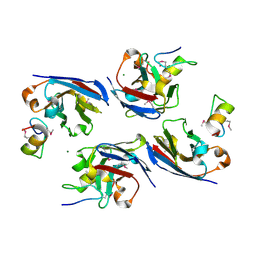 | | The complex of nanobody 6101 with BCL11A ZF6 | | Descriptor: | B-cell lymphoma/leukemia 11A, MAGNESIUM ION, Nanobody 6101, ... | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DTU
 
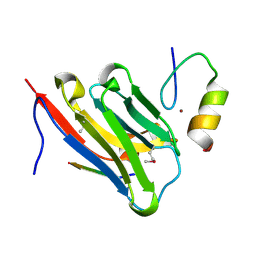 | | The complex of nanobody 5344N74D with BCL11A ZF6. | | Descriptor: | B-cell lymphoma/leukemia 11A, Nanobody 5344N74D, ZINC ION | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YLK
 
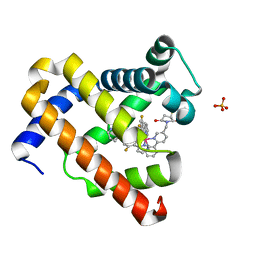 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
8DTM
 
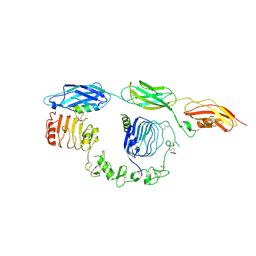 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | Descriptor: | Insulin mimetic peptide S597 component 2, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DU3
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with compound 21a | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4M)-6-bromo-4-(furan-2-yl)quinazolin-2-amine, Adenosine receptor A2a, ... | | Authors: | Shiriaeva, A, Stauch, B, Han, G.W, Cherezov, V. | | Deposit date: | 2022-07-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High ligand efficiency quinazoline compounds as novel A 2A adenosine receptor antagonists.
Eur.J.Med.Chem., 241, 2022
|
|
7YL1
 
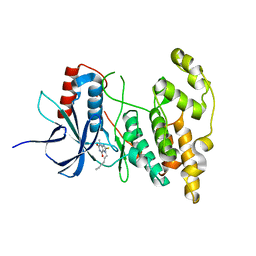 | |
7YKZ
 
 | | Cryo-EM structure of Drg1 hexamer in the planar state treated with ADP/AMPPNP/Diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
8DTL
 
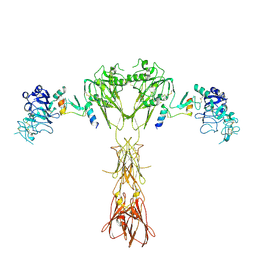 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8AHY
 
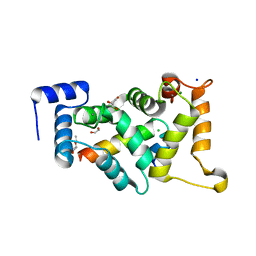 | |
