2I56
 
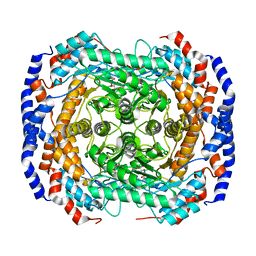 | | Crystal structure of L-Rhamnose Isomerase from Pseudomonas stutzeri with L-Rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
7LGJ
 
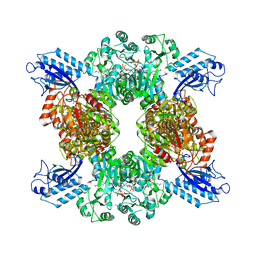 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 8x(Asp-Arg)-NH2 | | Descriptor: | 8x(Asp-Arg)-NH2, Cyanophycin synthase, MAGNESIUM ION, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
5LNH
 
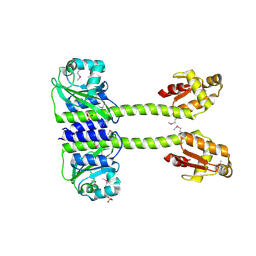 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
7E17
 
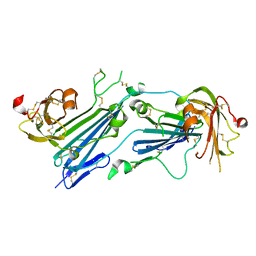 | | Structure of dimeric uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Cai, Y, Huang, M. | | Deposit date: | 2021-02-01 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
7C39
 
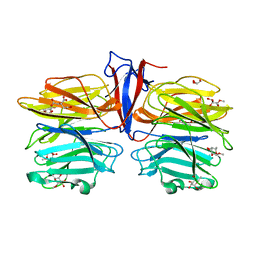 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with methylated L-fucose | | Descriptor: | AoflcA, CITRIC ACID, GLYCEROL, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
3J1W
 
 | |
7LME
 
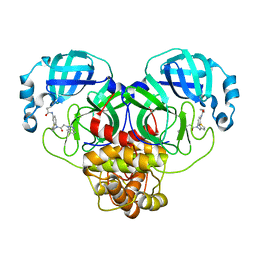 | | SARS-CoV-2 3CLPro in complex with N-[4-[[2-(benzotriazol-1-yl)acetyl]-(3-thienylmethyl)amino]phenyl]cyclopropanecarboxamide | | Descriptor: | 3C-like proteinase, ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | | Authors: | Goins, C.M, Arya, T, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
5LOK
 
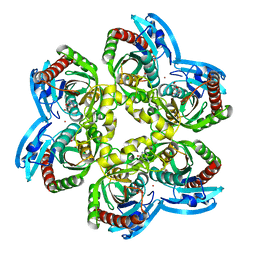 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Dontsova, M.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.109 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution
To Be Published
|
|
2HYB
 
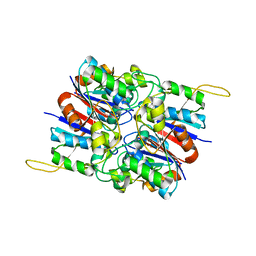 | | Crystal Structure of Hexameric DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexameric DsrEFH
To be Published
|
|
7LGU
 
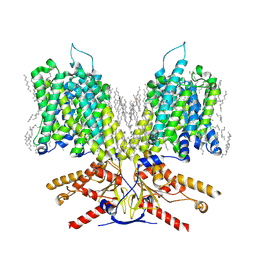 | |
7E7F
 
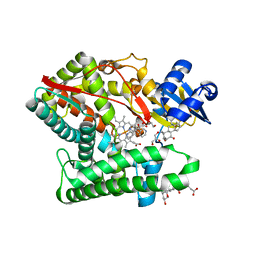 | | Human CYP11B1 mutant in complex with metyrapone | | Descriptor: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
3BGP
 
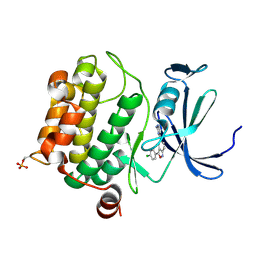 | | Human Pim-1 complexed with a benzoisoxazole inhibitor VX1 | | Descriptor: | 4-[3-(4-chlorophenyl)-2,1-benzisoxazol-5-yl]pyrimidin-2-amine, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Docking study yields four novel inhibitors of the protooncogene pim-1 kinase.
J.Med.Chem., 51, 2008
|
|
2I8U
 
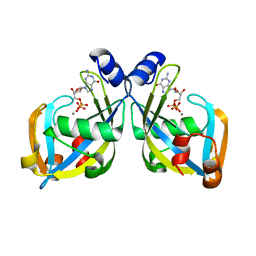 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
7EA2
 
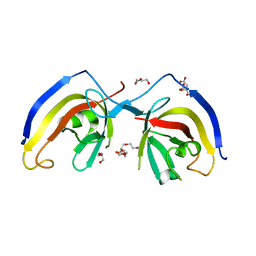 | | crystal structure of NAP1 FIR in complex with RB1CC1 Claw domain | | Descriptor: | 5-azacytidine-induced protein 2,RB1-inducible coiled-coil protein 1, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
5LQP
 
 | |
3K90
 
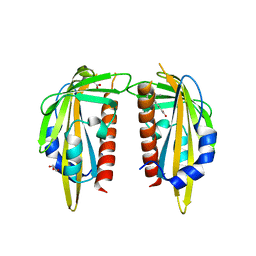 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2009-10-15 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
2ON6
 
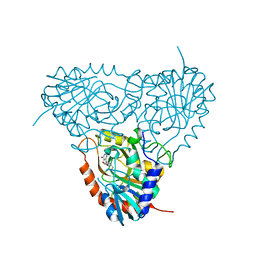 | | Crystal structure of human purine nucleoside phosphorylase mutant H257F with Imm-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Murkin, A.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Neighboring Group Participation in the Transition State of Human Purine Nucleoside Phosphorylase
Biochemistry, 46, 2007
|
|
7E9Y
 
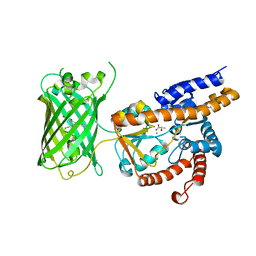 | | Crystal structure of eLACCO1 | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CALCIUM ION, Lactate-binding periplasmic protein TTHA0766,Lactate-binding periplasmic protein TTHA0766 | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J, Nasu, Y. | | Deposit date: | 2021-03-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A genetically encoded fluorescent biosensor for extracellular L-lactate.
Nat Commun, 12, 2021
|
|
2OM0
 
 | | Structure of human insulin in presence of urea at pH 6.5 | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Norrman, M, Schluckebier, G. | | Deposit date: | 2007-01-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic characterization of two novel crystal forms of human insulin induced by chaotropic agents and a shift in pH.
Bmc Struct.Biol., 7, 2007
|
|
5LVD
 
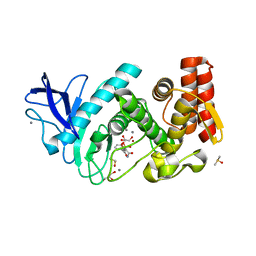 | | Thermolysin in complex with inhibitor (JC67) | | Descriptor: | (2~{S})-4-methyl-2-[[(2~{S})-3-oxidanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
2I1D
 
 | | DPC micelle-bound NMR structures of Tritrp1 | | Descriptor: | 13-mer from Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
3J3X
 
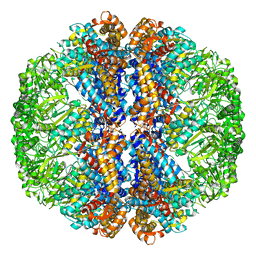 | |
2ONC
 
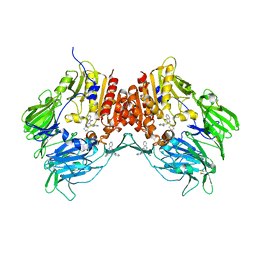 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
7DU4
 
 | |
2OOV
 
 | |
