5OJW
 
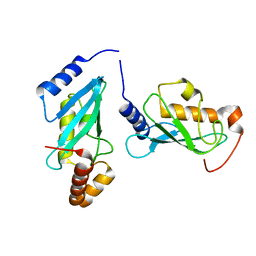 | | S. cerevisiae UBC13 - MMs2 complex | | Descriptor: | Ubiquitin-conjugating enzyme E2 13, Ubiquitin-conjugating enzyme variant MMS2 | | Authors: | Sharon, I, Rathi, R, Levin-Kravets, O, Attali, I, Prag, G. | | Deposit date: | 2017-07-24 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S. cerevisiae UBC13 - MMs2 complex
To Be Published
|
|
8DQM
 
 | |
8DQN
 
 | |
8EIP
 
 | |
8EIN
 
 | |
7TXV
 
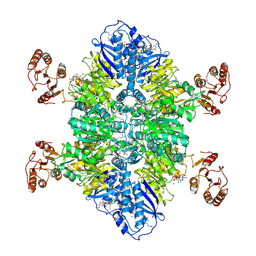 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q with ATP and 16x(Asp-Arg) | | Descriptor: | 16x(Asp-Arg), ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Nat Commun, 13, 2022
|
|
7TXU
 
 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) | | Descriptor: | 16x(Asp-Arg), ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Nat Commun, 13, 2022
|
|
7LGQ
 
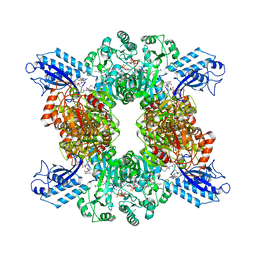 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 8x(Asp-Arg)-Asn | | Descriptor: | 8x(Asp-Arg)-Asn, ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7UQW
 
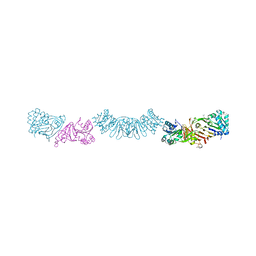 | | PCC6803 Cyanophycinase S132DAP covalently bound to cyanophycin dimer | | Descriptor: | (2~{S})-4-[[(2~{S})-5-[[azanyl($l^{4}-azanylidene)methyl]amino]-1-$l^{1}-oxidanyl-1-oxidanylidene-pentan-2-yl]amino]-2-$l^{2}-azanyl-4-oxidanylidene-butanoic acid, Cyanophycinase, FORMAMIDE, ... | | Authors: | Sharon, I, Schmeing, T.M. | | Deposit date: | 2022-04-20 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7LG5
 
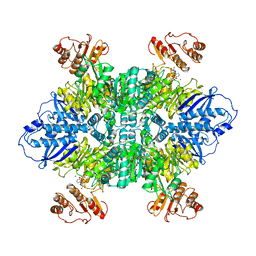 | |
7LGJ
 
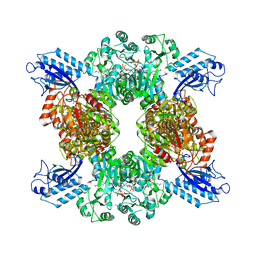 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 8x(Asp-Arg)-NH2 | | Descriptor: | 8x(Asp-Arg)-NH2, Cyanophycin synthase, MAGNESIUM ION, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7LGM
 
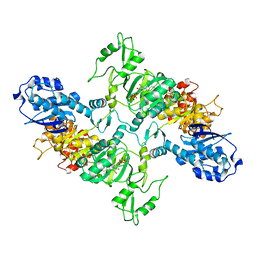 | | Cyanophycin synthetase from A. baylyi DSM587 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase | | Authors: | Sharon, I, Haque, A.S, Lahiri, I, Leschziner, A, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7LGN
 
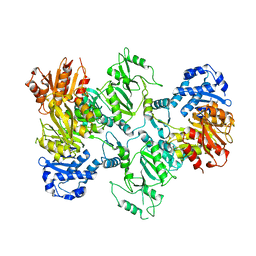 | |
7TA5
 
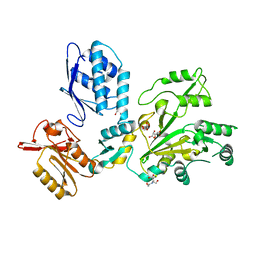 | |
7UQV
 
 | | Pseudobacteroides cellulosolvens pseudo-CphB | | Descriptor: | Cyanophycinase | | Authors: | Sharon, I, Schmeing, T.M. | | Deposit date: | 2022-04-20 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7LY4
 
 | | Cryo-EM structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC | | Descriptor: | BmdB, bacillamide NRPS, BmdC, ... | | Authors: | Sharon, I, Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7MLG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7MLF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
8FXI
 
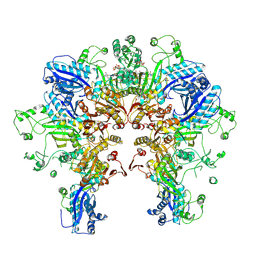 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
8FXH
 
 | |
7LY7
 
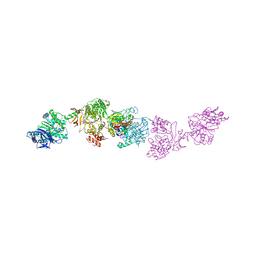 | | Crystal structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | 5'-{[(2R,3S)-3-amino-2-({2-[(N-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-4-sulfanylbutane-1-sulfonyl]amino}-5'-deoxyadenosine, BmdB, Bacillamide NRPS, ... | | Authors: | Fortinez, C.M, Sharon, I, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
