8PQ0
 
 | | Structure of human PARK7 in complex with GK16R | | Descriptor: | (3~{R})-3-(pent-4-ynylcarbamoyl)pyrrolidine-1-carboximidothioic acid, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PPW
 
 | | Structure of human PARK7 in complex with GK16S | | Descriptor: | (3~{S})-1-(iminomethyl)-~{N}-pent-4-ynyl-pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PPV
 
 | | Intermediate conformer of Pyrococcus abyssi DNA polymerase D (PolD) bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(*P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(G7P)P*(PST)P*(PST)P*(PST))-3'), DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPU
 
 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(C7R)P*(PST)P*(PST)P*(PST))-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPT
 
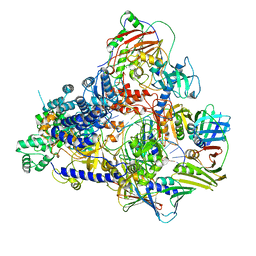 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing a mismatch | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*GP*CP*TP*TP*T)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPR
 
 | |
8PPL
 
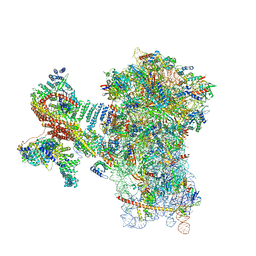 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPK
 
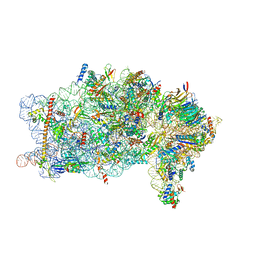 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PP7
 
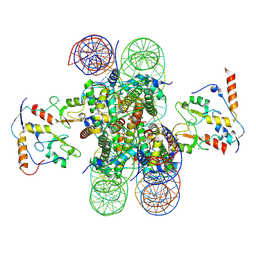 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 2024
|
|
8PP6
 
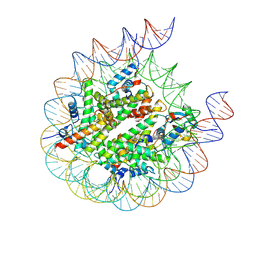 | |
8POH
 
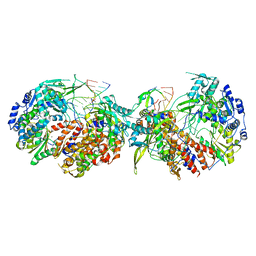 | |
8PNT
 
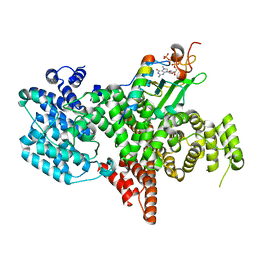 | | Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8PNQ
 
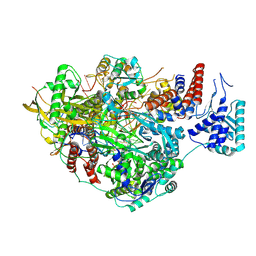 | |
8PNP
 
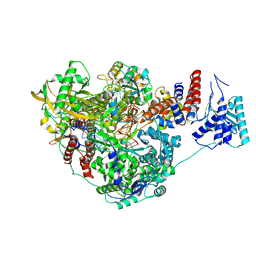 | |
8PMP
 
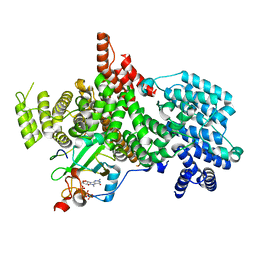 | | Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8PM0
 
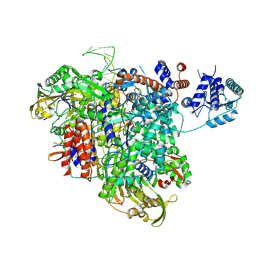 | |
8PLZ
 
 | | Cryo-EM structure of CAK in complex with inhibitor CT7030 | | Descriptor: | (3~{R},4~{R})-4-[[[7-[(2-methoxyphenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8PKI
 
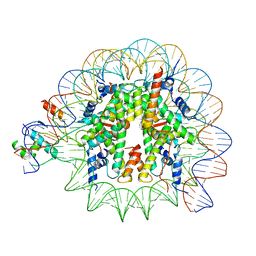 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PJJ
 
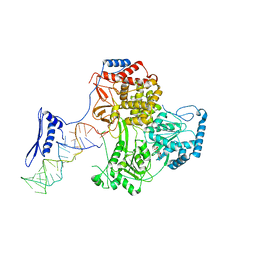 | |
8PJB
 
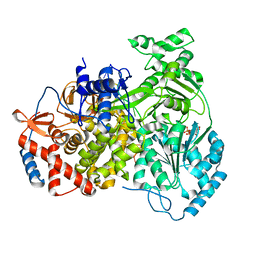 | |
8PEP
 
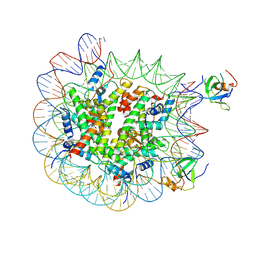 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PEO
 
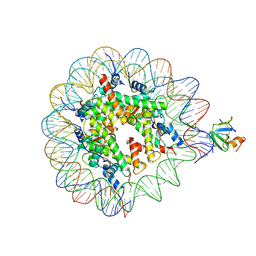 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | (2R)-2-amino-3-(2-dimethylaminoethylsulfanyl)propanoic acid, Histone H2A, Histone H2B 1.1, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PEG
 
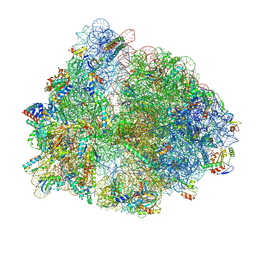 | |
8PC6
 
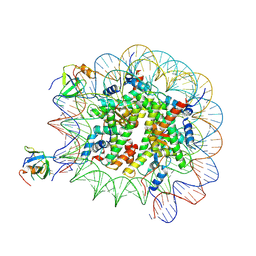 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PC5
 
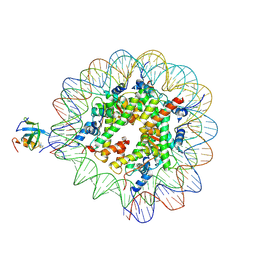 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
