6M3C
 
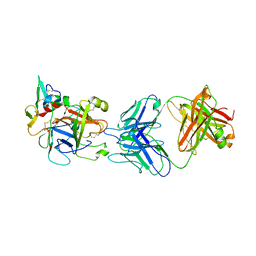 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
7MFG
 
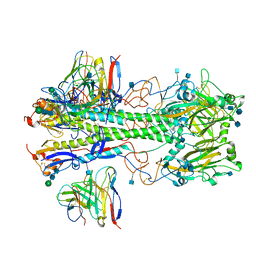 | | Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 310-030-1D06 Heavy, 310-030-1D06 Light, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-04-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
8E7W
 
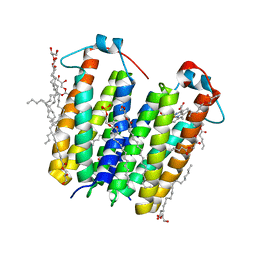 | | RsTSPO A139T with Heme | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6PFX
 
 | |
6UMG
 
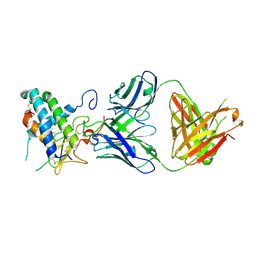 | | Crystal structure of erenumab Fab bound to the extracellular domain of CGRP receptor | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 1, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6ZV6
 
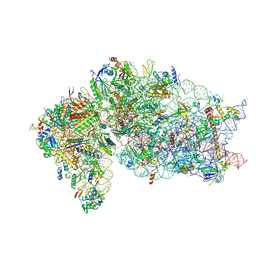 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
7M2K
 
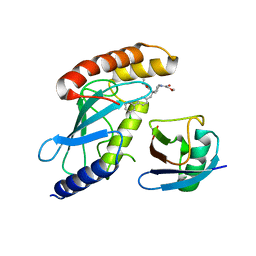 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
6M4S
 
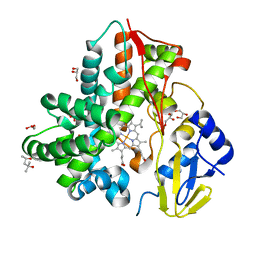 | | Crystal Structure Analysis of the cytochrome P450 CYP-Sb21 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cytochrome P450 hydroxylase sb21, ... | | Authors: | Li, F.W, Li, S.Y. | | Deposit date: | 2020-03-09 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana .
Synth Syst Biotechnol, 5, 2020
|
|
6PGN
 
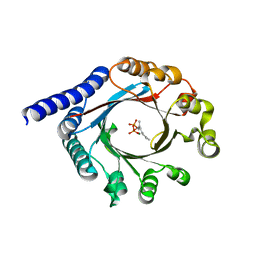 | | PagF single mutant with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, PagF | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in
Ribosomally Synthesized and Post-Translationally Modified Peptide
Prenyltransferases
J. Am. Chem. Soc., 140, 2018
|
|
6UML
 
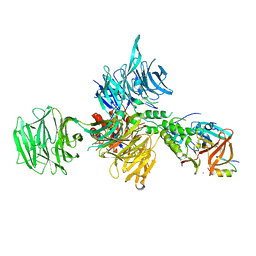 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7AC8
 
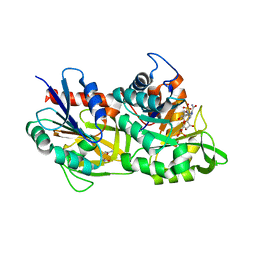 | |
8E0N
 
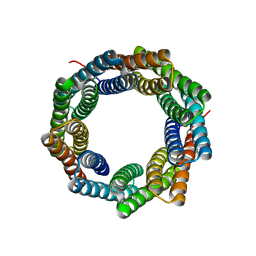 | |
7M4S
 
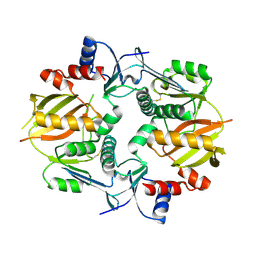 | |
6M2Q
 
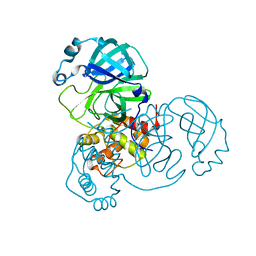 | | SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21) | | Descriptor: | 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
6M3B
 
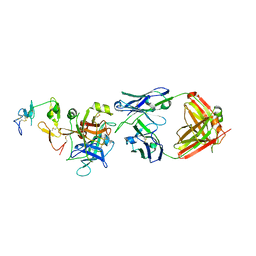 | | hAPC-c25k23 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, ... | | Authors: | Wang, X, Li, L, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
6ZOD
 
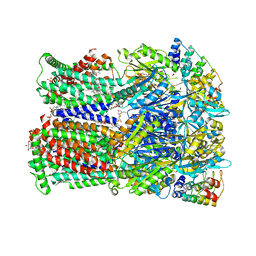 | | Fusidic acid binding to the allosteric deep transmembrane domain binding pocket, TM7/TM8 groove, and TM1/TM2 groove of the fully induced AcrB T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Oswald, C, Tam, H.K, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
5PVC
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 226) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
8E84
 
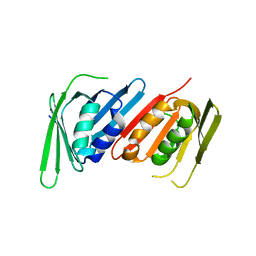 | | Human PCNA mutant- C148S | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Magrino, J, Page, B, Gaubitz, C, Kelch, B.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A thermosensitive PCNA allele underlies an ataxia-telangiectasia-like disorder.
J.Biol.Chem., 299, 2023
|
|
6P7B
 
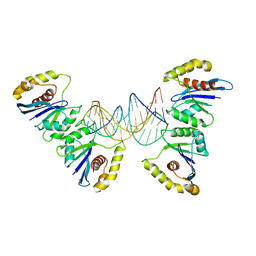 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
7MK1
 
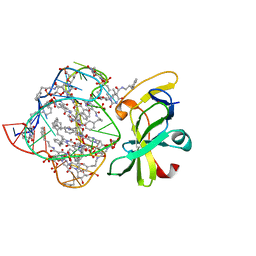 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
5PVS
 
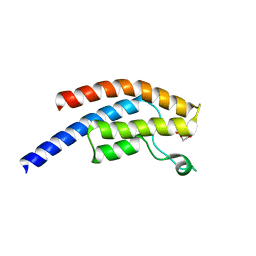 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 243) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6M3W
 
 | | Post-fusion structure of SARS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Fan, X, Cao, D, Zhang, X. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Nat Commun, 11, 2020
|
|
7MGK
 
 | |
6P7K
 
 | | Structure of HMG-CoA reductase from Burkholderia cenocepacia | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A reductase, ADENOSINE-5'-DIPHOSPHATE, COENZYME A | | Authors: | Walker, A.M, Peacock, R.B, Hicks, C.W, Dewing, S.M, Lewis, K.M, Abboud, J, Stewart, S.W.A, Kang, C, Watson, J.M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural and Functional Characterization of Dynamic Oligomerization in Burkholderia cenocepacia HMG-CoA Reductase.
Biochemistry, 58, 2019
|
|
6TZP
 
 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
