1RG2
 
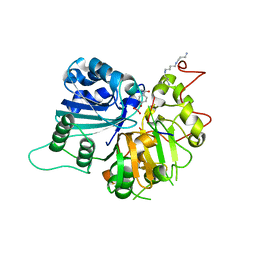 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTA | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*A)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
3H0G
 
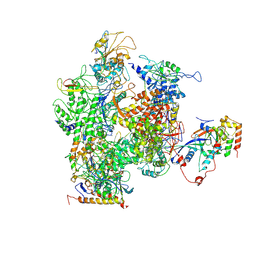 | | RNA Polymerase II from Schizosaccharomyces pombe | | Descriptor: | DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Spahr, H, Calero, G, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1R6U
 
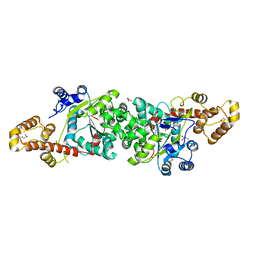 | | Crystal structure of an active fragment of human tryptophanyl-tRNA synthetase with cytokine activity | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Yang, X.-L, Otero, F.J, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase
Structure, 15, 2007
|
|
1AWD
 
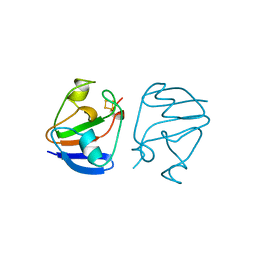 | | FERREDOXIN [2FE-2S] OXIDIZED FORM FROM CHLORELLA FUSCA | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Sheldrick, G.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure determination at 1.4 A resolution of ferredoxin from the green alga Chlorella fusca
Structure Fold.Des., 7, 1999
|
|
1UKH
 
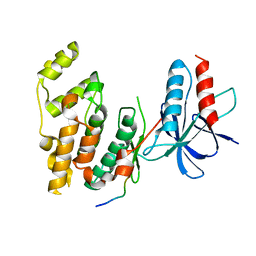 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, Mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
1ULZ
 
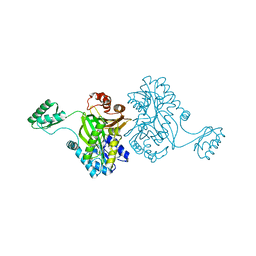 | | Crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase n-terminal domain | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Yong-Biao, J, Sueda, S, Kondo, H. | | Deposit date: | 2003-09-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UKI
 
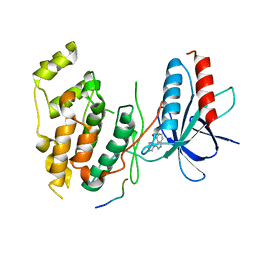 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
3LZM
 
 | |
1ROS
 
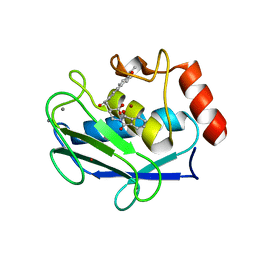 | | Crystal structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-[2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL)ETHYL]-4-(4'-ETHOXY-1,1'-BIPHENYL-4-YL)-4-OXOBUTANOIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J.Mol.Biol., 341, 2004
|
|
1RGU
 
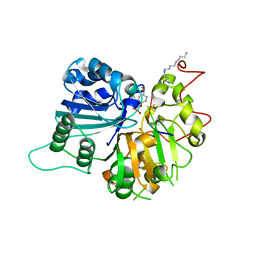 | | The crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTG | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*G)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1IWD
 
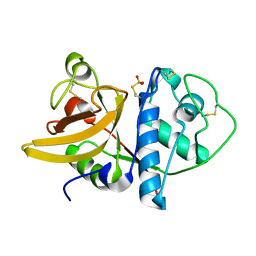 | |
1SIV
 
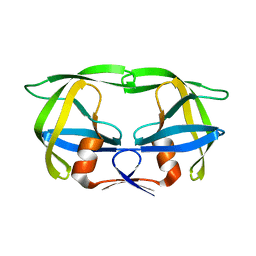 | |
1XMW
 
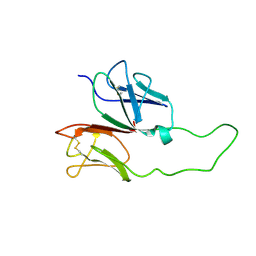 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | Descriptor: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S5O
 
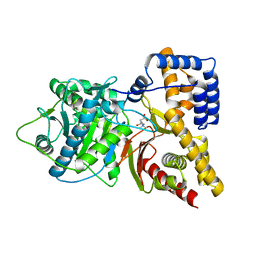 | | Structural and Mutational Characterization of L-carnitine Binding to Human carnitine Acetyltransferase | | Descriptor: | CARNITINE, carnitine acetyltransferase isoform 2 | | Authors: | Govindasamy, L, Kukar, T, Lian, W, Pedersen, B, Gu, Y, Agbandje-Mckenna, M, Jin, S, Mckenna, R, Wu, D. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational characterization of l-carnitine binding to human carnitine acetyltransferase.
J.Struct.Biol., 146, 2004
|
|
1WMA
 
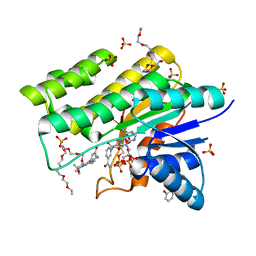 | | Crystal structure of human CBR1 in complex with Hydroxy-PP | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, ... | | Authors: | Rauh, D, Bateman, R, Shokat, K.M. | | Deposit date: | 2004-07-06 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An unbiased cell morphology-based screen for new, biologically active small molecules
Plos Biol., 3, 2005
|
|
1DST
 
 | |
2L3Y
 
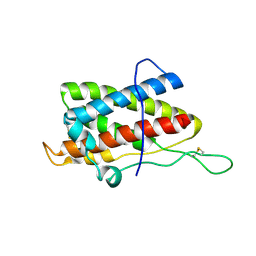 | | Solution structure of mouse IL-6 | | Descriptor: | Interleukin-6 | | Authors: | Veverka, V, Redpath, N.T, Carrington, B, Muskett, F.W, Taylor, R.J, Henry, A.J, Carr, M.D. | | Deposit date: | 2010-09-25 | | Release date: | 2011-09-28 | | Last modified: | 2013-01-02 | | Method: | SOLUTION NMR | | Cite: | Conservation of functional sites on interleukin-6 and implications for evolution of signaling complex assembly and therapeutic intervention.
J.Biol.Chem., 287, 2012
|
|
2LWP
 
 | |
2BBQ
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF POLYGLUTAMYL FOLATES BY THYMIDYLATE SYNTHASE | | Descriptor: | 10-PARPARGYL-5,8-DIDEAZAFOLATE-4-GLUTAMIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Kamb, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1992-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of polyglutamyl folates by thymidylate synthase.
Biochemistry, 31, 1992
|
|
1ESO
 
 | | MONOMERIC CU,ZN SUPEROXIDE DISMUTASE FROM ESCHERICHIA COLI | | Descriptor: | COPPER (II) ION, CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Pesce, A, Capasso, C, Battistoni, A, Folcarelli, S, Rotilio, G, Desideri, A, Bolognesi, M. | | Deposit date: | 1997-06-27 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique structural features of the monomeric Cu,Zn superoxide dismutase from Escherichia coli, revealed by X-ray crystallography.
J.Mol.Biol., 274, 1997
|
|
2LSK
 
 | | C-terminal domain of human REV1 in complex with DNA-polymerase H (eta) | | Descriptor: | DNA polymerase eta, DNA repair protein REV1 | | Authors: | Pozhidaeva, A, Pustovalova, Y, Bezsonova, I, Korzhnev, D. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta.
Biochemistry, 51, 2012
|
|
2KG0
 
 | |
2KFY
 
 | |
2JJJ
 
 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
2JJI
 
 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
