8OOY
 
 | | Pol I bound to extended and displaced DNA section - open conformation | | Descriptor: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OQ9
 
 | | Crystal structure of the titin domain Fn3-56 | | Descriptor: | CHLORIDE ION, Titin, ZINC ION | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
5HRO
 
 | |
8OMW
 
 | | Crystal structure of the titin domain Fn3-20 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8ORL
 
 | |
8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OTY
 
 | | Crystal structure of the titin domain Fn3-90 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OT5
 
 | | Crystal structure of the titin domain Fn3-85 | | Descriptor: | CHLORIDE ION, SODIUM ION, Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
5HVF
 
 | | Crystal Structure of Thrombin-activatable Fibrinolysis Inhibitor in Complex with an Inhibitory Nanobody (VHH-i83) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Carboxypeptidase B2, ... | | Authors: | Zhou, X, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Elucidation of the molecular mechanisms of two nanobodies that inhibit thrombin-activatable fibrinolysis inhibitor activation and activated thrombin-activatable fibrinolysis inhibitor activity.
J.Thromb.Haemost., 14, 2016
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
8OZE
 
 | | cryoEM structure of SPARTA complex dimer high resolution | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*G)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OR9
 
 | |
8OZI
 
 | | cryoEM structure of SPARTA complex pre-NAD cleavage | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8P3U
 
 | |
8OUW
 
 | | Cryo-EM structure of CMG helicase bound to TIM-1/TIPN-1 and homodimeric DNSN-1 on fork DNA (Caenorhabditis elegans) | | Descriptor: | Cell division control protein 45 homolog, DNA Lagging Strand Template, DNA Leading Strand Template, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Labib, K.P.M. | | Deposit date: | 2023-04-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans.
Science, 381, 2023
|
|
8OZG
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-1 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4WEF
 
 | | Structure of the Hemagglutinin-neuraminidase from Human parainfluenza virus type III: complex with difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
8P4B
 
 | | Structural insights into human co-transcriptional capping - structure 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*AP*GP*CP*GP*GP*CP*GP*GP*AP*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*AP*GP*CP*TP*G)-3'), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8OZD
 
 | | cryoEM structure of SPARTA complex dimer-3 | | Descriptor: | DNA (16-MER), Piwi domain-containing protein, RNA (18-MER), ... | | Authors: | Ekundayo, B, Ni, D.C, Lu, X.H, Stahlberg, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZ6
 
 | | cryoEM structure of SPARTA complex ligand-free | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8ORA
 
 | | Human holo aromatic L-amino acid decarboxylase (AADC) external aldimine with L-Dopa methylester | | Descriptor: | Dopa decarboxylase (Aromatic L-amino acid decarboxylase), PYRIDOXAL-5'-PHOSPHATE, TETRAETHYLENE GLYCOL, ... | | Authors: | Bisello, G, Perduca, M, Bertoldi, M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: Implication in aromatic amino acid decarboxylase deficiency.
Protein Sci., 32, 2023
|
|
8OX0
 
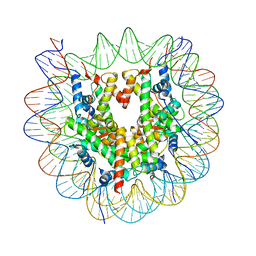 | | Structure of apo telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
8EIW
 
 | | Cobalt(II)-substituted Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
