8VEY
 
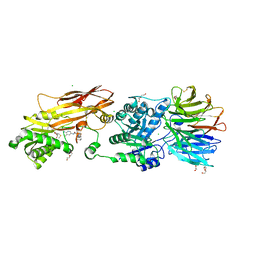 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG908 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEU
 
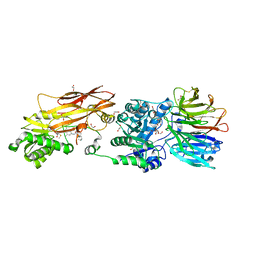 | |
8VET
 
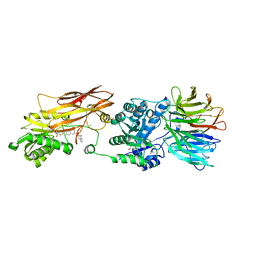 | |
8VEX
 
 | |
6CKC
 
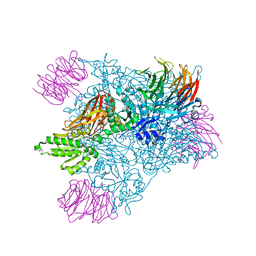 | | Structure of PRMT5:MEP50 in complex with LLY-283, a potent and selective inhibitor of PRMT5, with antitumor activity | | Descriptor: | 7-[(5R)-5-C-phenyl-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Antonysamy, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity.
ACS Med Chem Lett, 9, 2018
|
|
6ZLW
 
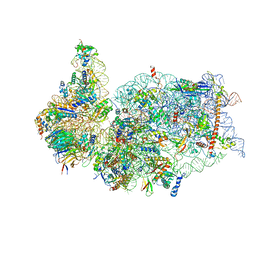 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMT
 
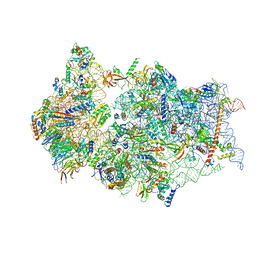 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZON
 
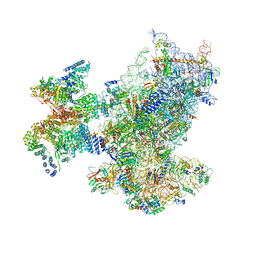 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
8G1U
 
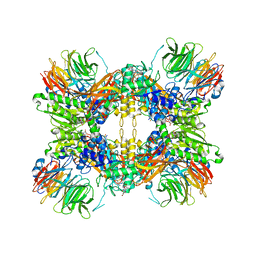 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
2RKJ
 
 | | Cocrystal structure of a tyrosyl-tRNA synthetase splicing factor with a group I intron RNA | | Descriptor: | RNA (238-MER), RNA (5'-R(P*GP*CP*UP*U)-3'), Tyrosyl-tRNA synthetase | | Authors: | Paukstelis, P.J, Chen, J.-H, Chase, E, Lambowitz, A.M, Golden, B.L. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
Nature, 451, 2008
|
|
8VEO
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
1QF6
 
 | | STRUCTURE OF E. COLI THREONYL-TRNA SYNTHETASE COMPLEXED WITH ITS COGNATE TRNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, THREONINE TRNA, THREONYL-TRNA SYNTHETASE, ... | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 1999-04-06 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site
Cell(Cambridge,Mass.), 97, 1999
|
|
1WZ2
 
 | |
8QDV
 
 | |
7M05
 
 | |
8PPL
 
 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
2BTE
 
 | | Thermus thermophilus Leucyl-tRNA synthetase complexed with a tRNAleu transcript in the post-editing conformation and a post- transfer editing substrate analogue | | Descriptor: | 2'-AMINO-2'-DEOXYADENOSINE, AMINOACYL-TRNA SYNTHETASE, LEUCINE, ... | | Authors: | Cusack, S, Tukalo, M, Yaremchuk, A, Fukunaga, R, Yokoyama, S. | | Deposit date: | 2005-05-31 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Leucyl-tRNA Synthetase Complexed with tRNA(Leu) in the Post-Transfer- Editing Conformation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7ZV2
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZVL
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZUU
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZUQ
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZUY
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZUP
 
 | | Human PRMT5:MEP50 structure with Fragment (Example 18) and MTA Bound | | Descriptor: | 3-ethylimidazo[4,5-b]pyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
