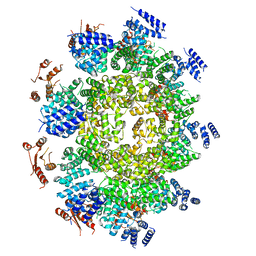
6WPK
 
 | |
6X25
 
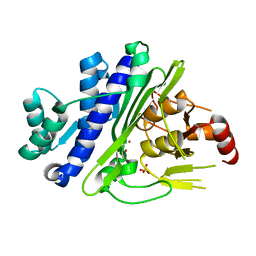 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6UJS
 
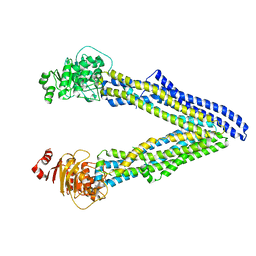 | | P-glycoprotein mutant-F728A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UML
 
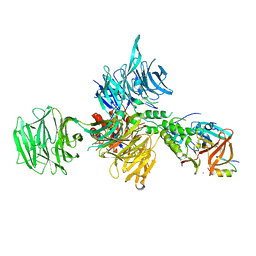 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3E7I
 
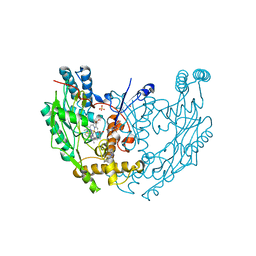 | | Structure of murine inos oxygenase domain with inhibitor AR-C94864 | | Descriptor: | (2R)-5-FLUORO-2-(2-THIENYL)-1,2-DIHYDROQUINAZOLIN-4-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E46
 
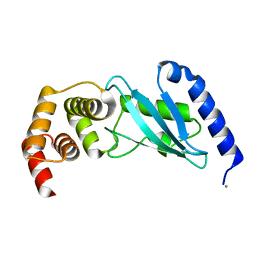 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington interacting protein 2) M172A mutant | | Descriptor: | CALCIUM ION, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Hughes, R.C, Wilson, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-08-09 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3E7T
 
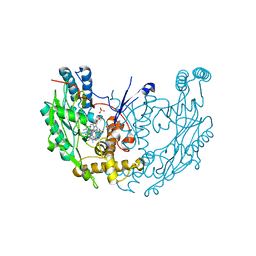 | | Structure of murine iNOS oxygenase domain with inhibitor AR-C102222 | | Descriptor: | 1-(6-CYANO-3-PYRIDYLCARBONYL)-5',8'-DIFLUOROSPIRO[PIPERIDINE-4,2'(1'H)-QUINAZOLINE]-4'-AMINE, 7,8-DIHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
6UD1
 
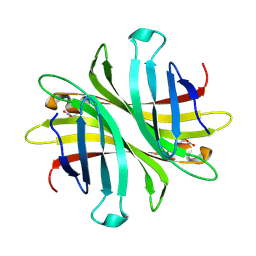 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | CHLORIDE ION, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
3E6L
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C132283 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-CHLOROPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EDF
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase, ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3E67
 
 | | Murine inos dimer with inhibitor 4-MAP bound | | Descriptor: | 4-METHYLPYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
6UO8
 
 | | Human metabotropic GABA(B) receptor bound to agonist SKF97541 and positive allosteric modulator GS39783 | | Descriptor: | (R)-(3-aminopropyl)methylphosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2019-10-14 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
6UOX
 
 | | Structure of itraconazole-bound NPC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-bromo-4-{4-[4-({(2R,4S)-2-(2,4-dichlorophenyl)-2-[(1H-1,2,4-triazol-1-yl)methyl]-1,3-dioxolan-4-yl}methoxy)phenyl]piperazin-1-yl}phenyl)-2-[(2S)-butan-2-yl]-2,4-dihydro-3H-1,2,4-triazol-3-one, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structural basis for itraconazole-mediated NPC1 inhibition.
Nat Commun, 11, 2020
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WDD
 
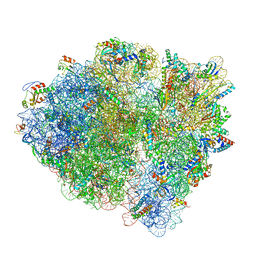 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WD5
 
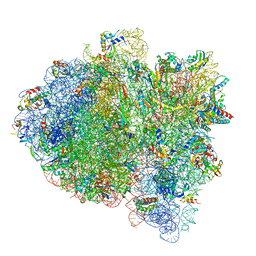 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-C1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WEA
 
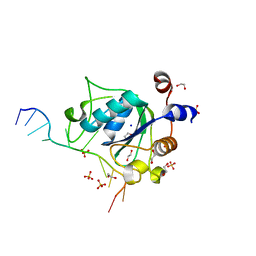 | | YTH domain of human YTHDC1 with a 10mer Oligo Containing N6mA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*GP*(6MA)P*CP*TP*TP*C)-3'), SODIUM ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
3FFK
 
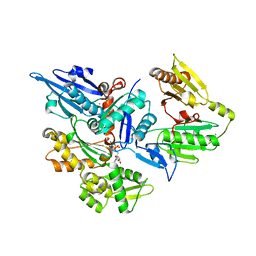 | | Crystal structure of human Gelsolin domains G1-G3 bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, actin, ... | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Burtnick, L.D. | | Deposit date: | 2008-12-03 | | Release date: | 2009-10-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6WDM
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3FJW
 
 | | Crystal Structure Analysis of Fungal Versatile Peroxidase from Pleurotus eryngii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Piontek, K, Martinez, A.T, Choinowski, T, Plattner, D.A. | | Deposit date: | 2008-12-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Site-directed Mutagenesis Study of Versatile Peroxidase Oxidizing both Mn(II) and Aromatic Substrates
To be Published
|
|
6WKJ
 
 | | Crystal structure of pentalenene synthase mutant F76H complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
3FM1
 
 | | Crystal Structure Analysis of Fungal Versatile Peroxidase from Pleurotus eryngii | | Descriptor: | CALCIUM ION, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Piontek, K, Martinez, A.T, Choinowski, T, Plattner, D.A. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Site-directed Mutagenesis Study of Versatile Peroxidase Oxidizing both Mn(II) and Aromatic Substrates
To be Published
|
|
6WOT
 
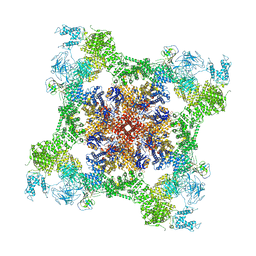 | | Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
6WP6
 
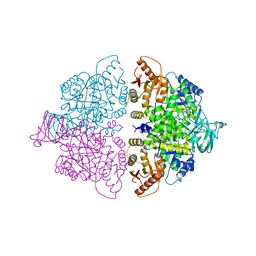 | | Pyruvate Kinase M2 mutant-S37E K433E | | Descriptor: | GLYCEROL, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
