6VZ2
 
 | | Escherichia coli transcription-translation complex D1 (TTC-D1) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VJ9
 
 | |
3COA
 
 | | Crystal Structure of FoxO1 DBD Bound to IRE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DCP*DAP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DGP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DG)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
6W1V
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WDC
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure IV-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WE8
 
 | | YTH domain of human YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
3CQK
 
 | | Crystal Structure of L-xylulose-5-phosphate 3-epimerase UlaE (form B) complex with Zn2+ and sulfate | | Descriptor: | L-ribulose-5-phosphate 3-epimerase ulaE, SULFATE ION, ZINC ION | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of L-xylulose-5-Phosphate 3-epimerase (UlaE) from the anaerobic L-ascorbate utilization pathway of Escherichia coli: identification of a novel phosphate binding motif within a TIM barrel fold.
J.Bacteriol., 190, 2008
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WD4
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3CRT
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
6WIV
 
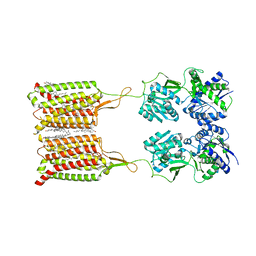 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
6WJC
 
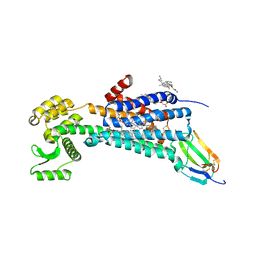 | | Muscarinic acetylcholine receptor 1 - muscarinic toxin 7 complex | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, ACETAMIDE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Kobilka, B.K. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and selectivity engineering of the M1muscarinic receptor toxin complex.
Science, 369, 2020
|
|
6WDL
 
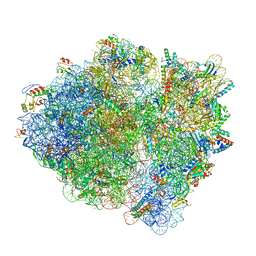 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-B1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3CO6
 
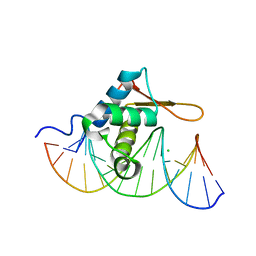 | | Crystal Structure of FoxO1 DBD Bound to DBE1 DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3CXX
 
 | |
6WD0
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure I-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3COT
 
 | | Crystal structure of human liver delta(4)-3-ketosteroid 5beta-reductase (akr1d1) in complex with progesterone and nadp. Resolution: 2.03 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
6WDA
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WDI
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure IV-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3CQJ
 
 | |
6WF2
 
 | | Crystal structure of mouse SCD1 with a diiron center | | Descriptor: | Acyl-CoA desaturase 1, FE (III) ION, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shen, J, Zhou, M. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase.
J.Mol.Biol., 432, 2020
|
|
6WB2
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WBL
 
 | |
6VTK
 
 | |
6VUM
 
 | |
