8AYM
 
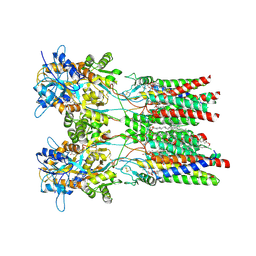 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-55511118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, ... | | Authors: | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
5O9A
 
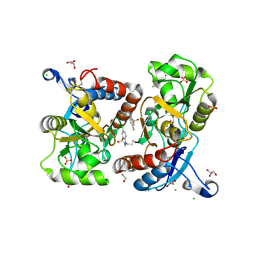 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504Y-N775S) in complex with glutamate and BPAM121 at 1.78 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Rovinskaja, K, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
8AYO
 
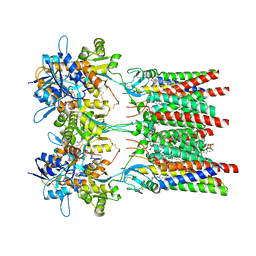 | | Open state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-61432059 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 5-[2-(4-fluorophenyl)-7-(4-oxidanylpiperidin-1-yl)pyrazolo[1,5-c]pyrimidin-3-yl]-1,3-dihydroindol-2-one, ... | | Authors: | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
5OEW
 
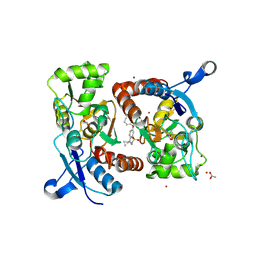 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with glutamate and positive allosteric modulator BPAM538 | | Descriptor: | 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
4Z0I
 
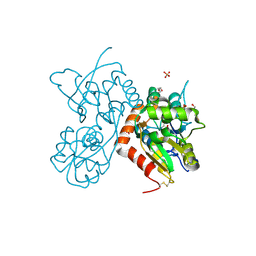 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
1SD3
 
 | |
1TXF
 
 | |
1SYI
 
 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1TT1
 
 | |
4ZDM
 
 | | Pleurobrachia bachei iGluR3 LBD Glycine Complex | | Descriptor: | GLYCINE, Glutamate receptor kainate-like protein, SODIUM ION, ... | | Authors: | Grey, R.J, Mayer, M.L. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CC2
 
 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
1S50
 
 | |
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
5CBS
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid at 1.8A resolution | | Descriptor: | (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
5BUU
 
 | | Crystal structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM-321 at 2.07 A resolution | | Descriptor: | (3R)-7-chloro-2,3,4-trimethyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, 1,2-ETHANEDIOL, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Tapken, D, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors.
Acs Chem Neurosci, 7, 2016
|
|
5CMK
 
 | | Crystal structure of the GluK2EM LBD dimer assembly complex with glutamate and LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Chittori, S, Mayer, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5CMM
 
 | |
5CBR
 
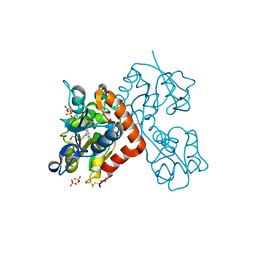 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(3,4-dichloro-5-(5-hydroxypyridin-3-yl)phenyl)propanoic acid at 2.0A resolution | | Descriptor: | 3,4-dichloro-5-(5-hydroxypyridin-3-yl)-L-phenylalanine, ACETATE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
5DEX
 
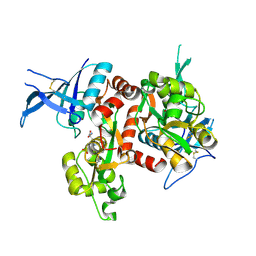 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
1S9T
 
 | |
1S7Y
 
 | |
1VSO
 
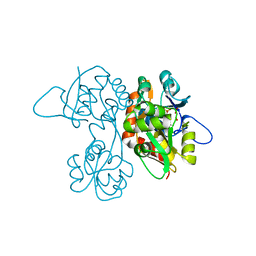 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
6WHY
 
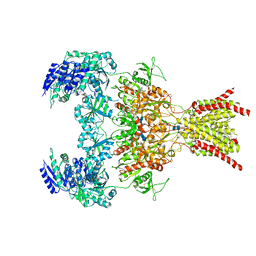 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
5KUF
 
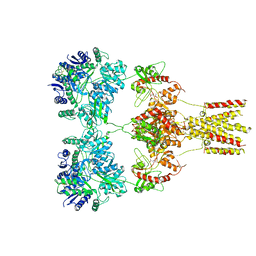 | | GluK2EM with 2S,4R-4-methylglutamate | | Descriptor: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5L1F
 
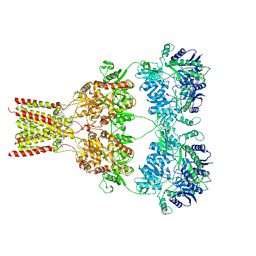 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor Perampanel | | Descriptor: | 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
