1XGB
 
 | |
1XGA
 
 | |
7EGR
 
 | | Co-crystal structure of Ac-AChBPP in complex with RgIA | | Descriptor: | MAGNESIUM ION, RgIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Pan, S, Fan, Y.X, Xue, Y, Zhu, X.P, Luo, S.L. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Co-crystal structure of Ac-AChBPP in complex with RgIA
To Be Published
|
|
8JPD
 
 | | Focused refinement structure of GRK2 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | Beta-adrenergic receptor kinase 1, STAUROSPORINE | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
1FF4
 
 | |
7EW2
 
 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
1MTQ
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID BY NMR SPECTROSCOPY | | Descriptor: | alpha-conotoxin GID | | Authors: | Nicke, A, Loughnan, M.L, Millard, E.L, Alewood, P.F, Adams, D.J, Daly, N.L, Craik, D.J, Lewis, R.J. | | Deposit date: | 2002-09-22 | | Release date: | 2003-02-11 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Isolation, Structure, and Activity of GID, a Novel alpha 4/7-Conotoxin with an Extended N-terminal Sequence
J.BIOL.CHEM., 278, 2003
|
|
5TVH
 
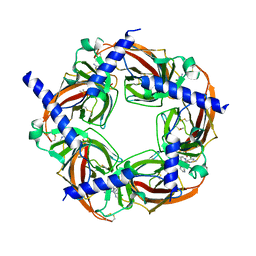 | | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 8.0 spacegroup P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N~4~,N~4~-bis[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, DIMETHYL SULFOXIDE, ... | | Authors: | Camacho-Hernandez, G.A, Kaczanowska, K, Harel, M, Finn, M.G, Taylor, P.W. | | Deposit date: | 2016-11-08 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121
To Be Published
|
|
6M4X
 
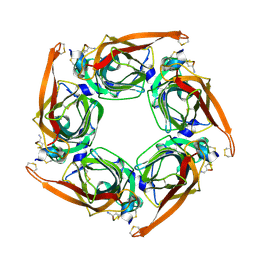 | |
6M4Z
 
 | |
3JAV
 
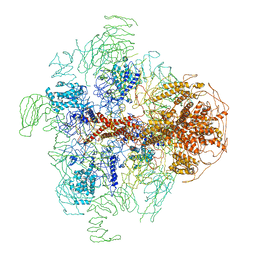 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
8YKY
 
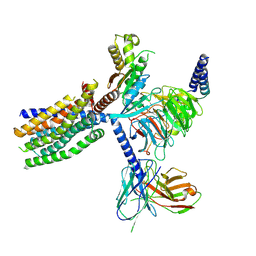 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQP
 
 | | Structure of human class T GPCR TAS2R14-Gustducin complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
7LQS
 
 | | Structure of truncated conotoxin CIC | | Descriptor: | Alpha-conotoxin CIC | | Authors: | Evans, E.R.J, Daly, N.L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Structural and Pharmacological Characterizations of CIC, a Novel alpha-Conotoxin with an Extended N-Terminal Tail.
Mar Drugs, 19, 2021
|
|
8EIT
 
 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJK
 
 | | Structure of FFAR1-Gq complex bound to TAK-875 in a lipid nanodisc | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJC
 
 | | Structure of FFAR1-Gq complex bound to TAK-875 | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6V0B
 
 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8YUT
 
 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | Descriptor: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8SXP
 
 | |
3V64
 
 | | Crystal Structure of agrin and LRP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 4, ... | | Authors: | Zong, Y, Zhang, B, Gu, S, Lee, K, Zhou, J, Yao, G, Figueiedo, D, Perry, K, Mei, L, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
3V65
 
 | | Crystal structure of agrin and LRP4 complex | | Descriptor: | Agrin, CALCIUM ION, Low-density lipoprotein receptor-related protein 4 | | Authors: | Zong, Y, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
8JD3
 
 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
