5Y7H
 
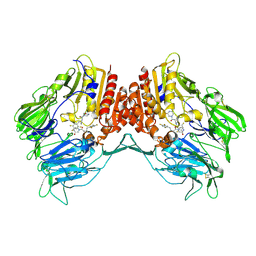 | | Crystal structure of human DPP4 in complex with inhibitor3 | | Descriptor: | (R)-4-(3-amino-4-(2,4,5-trifluorophenyl)butanoyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5G46
 
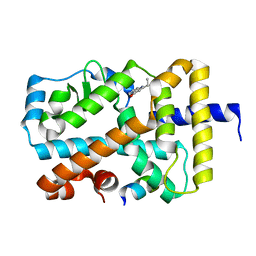 | | Ligand complex of RORg LBD | | Descriptor: | 2-ETHYL-4(1H)-QUINOLINONE, NUCLEAR RECEPTOR ROR-GAMMA, RORG, ... | | Authors: | Xue, Y, Guo, H, Hillertz, P. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment Screening of Rorgammat Using Cocktail Crystallography: Identification of Simultaneous Binding of Multiple Fragments.
Chemmedchem, 11, 2016
|
|
8GHP
 
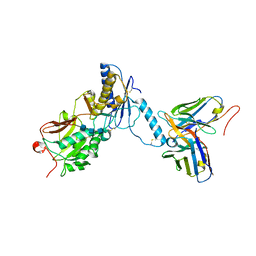 | | GUCY2C-ECD bound to anti-GUCY2C-scFv antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Guanylyl cyclase C, anti-GUCY2C-scFv antibody heavy chain, ... | | Authors: | Mosyak, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Molecular insights into recognition of GUCY2C by T-cell engaging bispecific antibody anti-GUCY2CxCD3.
Sci Rep, 13, 2023
|
|
6UZW
 
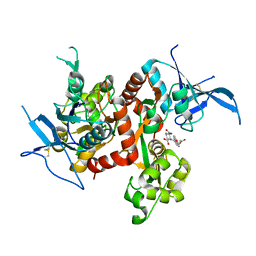 | | Crystal structure of GLUN1/GLUN2A ligand-binding domain in complex with glycine and UBP791 | | Descriptor: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Wang, J.X, Furukawa, H. | | Deposit date: | 2019-11-15 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6V02
 
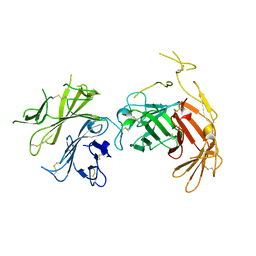 | | N-terminal 5 domains of CI-MPR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
5TZR
 
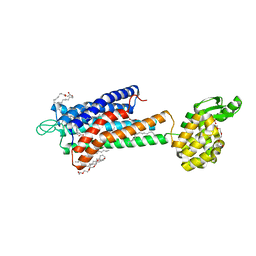 | | GPR40 in complex with partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5GHV
 
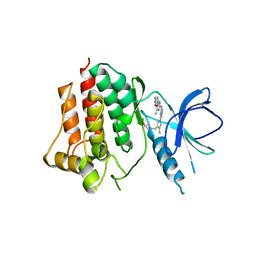 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4IU5
 
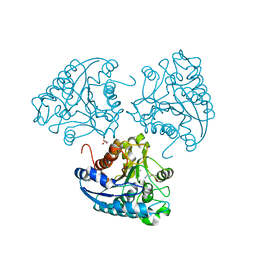 | | Crystal structure of Leishmania mexicana arginase in complex with catalytic product L-ornithine | | Descriptor: | Arginase, GLYCEROL, L-ornithine, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
5U3F
 
 | | Structure of Mycobacterium tuberculosis IlvE, a branched-chain amino acid transaminase, in complex with D-cycloserine derivative | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Branched-chain-amino-acid aminotransferase | | Authors: | Favrot, L, Amorim Franco, T.M, Blanchard, J.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Mechanism-Based Inhibition of the Mycobacterium tuberculosis Branched-Chain Aminotransferase by d- and l-Cycloserine.
ACS Chem. Biol., 12, 2017
|
|
8GHO
 
 | | GUCY2C-peptide bound to anti-GUCY2C-scFv antibody | | Descriptor: | Guanylyl cyclase C peptide, anti-GUCY2C-scFv antibody heavy chain, anti-GUCY2C-scFv antibody light chain | | Authors: | Mosyak, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into recognition of GUCY2C by T-cell engaging bispecific antibody anti-GUCY2CxCD3.
Sci Rep, 13, 2023
|
|
7SVP
 
 | |
5U58
 
 | | Psf4 in complex with Fe2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, FE (III) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
4ISA
 
 | | Crystal Structure of the Escherichia coli LpxC/BB-78485 complex | | Descriptor: | (2R)-N-hydroxy-3-naphthalen-2-yl-2-[(naphthalen-2-ylsulfonyl)amino]propanamide, (4S,5S)-1,2-DITHIANE-4,5-DIOL, FORMIC ACID, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4IS8
 
 | |
4IU0
 
 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, GLYCEROL, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
5UFJ
 
 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | Descriptor: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
7T1D
 
 | | Human SIRT2 in complex with small molecule 359 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2,4-dimethyl-1H-imidazol-1-yl)-2-(5-{[4-(1H-pyrazol-1-yl)phenyl]methyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Kulp, J.L, Remiszewski, S, Todd, M, Chiang, L.W. | | Deposit date: | 2021-12-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric inhibitor of sirtuin 2 deacetylase activity exhibits broad-spectrum antiviral activity.
J.Clin.Invest., 133, 2023
|
|
7ST5
 
 | | Structure of Fab CC-95251 in complex with SIRP-alpha | | Descriptor: | 1,2-ETHANEDIOL, Fab CC-95251 anti-SIRP-alpha heavy chain, Fab CC-95251 anti-SIRP-alpha light chain, ... | | Authors: | Fenalti, G. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Preclinical Activity of BMS-986351, an Antibody to SIRP alpha That Enhances Macrophage-mediated Tumor Phagocytosis When Combined with Opsonizing Antibodies.
Cancer Res Commun, 4, 2024
|
|
8GEM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethanamine | | Descriptor: | N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GDM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with {[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}(methyl)[1-(thiophen-2-yl)ethyl]amine | | Descriptor: | (1S)-N-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-N-methyl-1-(thiophen-2-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
5U94
 
 | | Crystal structure of the Mycobacterium tuberculosis PASTA kinase PknB in complex with the potential theraputic kinase inhibitor GSK690693. | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3 -yn-2-ol, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wlodarchak, N, Satyshur, K, Striker, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Silico Screen and Structural Analysis Identifies Bacterial Kinase Inhibitors which Act with beta-Lactams To Inhibit Mycobacterial Growth.
Mol. Pharm., 15, 2018
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
5Y9K
 
 | | Structure of the belimumab Fab fragment | | Descriptor: | belimumab heavy chain, belimumab light chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
5GPG
 
 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
4J28
 
 | | Crystal structure of a gh29 alpha-l-fucosidase gh29 from bacteroides thetaiotaomicron in complex with a 5-membered iminocyclitol inhibitor | | Descriptor: | (2S,3S,4R,5S)-2-(1H-benzimidazol-2-yl)-5-methylpyrrolidine-3,4-diol, Alpha-L-fucosidase, GLYCEROL, ... | | Authors: | Wright, D.W. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Three dimensional structure of a bacterial alpha-l-fucosidase with a 5-membered iminocyclitol inhibitor.
Bioorg.Med.Chem., 21, 2013
|
|
