5JFK
 
 | | Crystal structure of a TDR receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR | | Authors: | Li, Z, Xu, G. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Crystal structure of a TDR receptor
To Be Published
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5J5O
 
 | | Translation initiation factor 4E in complex with m7GppppG mRNA 5' cap analog | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]-7-methylguanosine, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
5N2S
 
 | | Crystal structure of stabilized A1 receptor in complex with PSB36 at 3.3A resolution | | Descriptor: | 1-butyl-3-(3-oxidanylpropyl)-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, SULFATE ION, Soluble cytochrome b562,Adenosine receptor A1 | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
5NP6
 
 | | 70S structure prior to bypassing | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Klimova, M, Zamora, M, Gil-Carton, D, Rodnina, M, Valle, M. | | Deposit date: | 2017-04-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome rearrangements at the onset of translational bypassing.
Sci Adv, 3, 2017
|
|
5JA3
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta- NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-b iphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hajian, B, Anderson, A.C. | | Deposit date: | 2016-04-11 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Propargyl-Linked Antifolates Are Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Plos One, 11, 2016
|
|
5MZP
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CAFFEINE, ... | | Authors: | Cheng, K.Y.R, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5KLI
 
 | | Rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5KOY
 
 | | Mouse pgp 34 linker deleted bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KO2
 
 | | Mouse pgp 34 linker deleted mutant Hg derivative | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
5LFR
 
 | |
5LG6
 
 | | Structure of the deglycosylated porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ZINC ION | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-07-06 | | Release date: | 2017-04-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5KQR
 
 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine | | Descriptor: | CHLORIDE ION, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Jain, R, Coloma, J, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
5KJ2
 
 | | The novel p300/CBP inhibitor A-485 uncovers a unique mechanism of action to target AR in castrate resistant prostate cancer | | Descriptor: | Histone acetyltransferase p300, N-[(4-fluorophenyl)methyl]-2-{(1R)-5-[(methylcarbamoyl)amino]-2',4'-dioxo-2,3-dihydro-3'H-spiro[indene-1,5'-[1,3]oxazolidin]-3'-yl}-N-[(2S)-1,1,1-trifluoropropan-2-yl]acetamide, SODIUM ION | | Authors: | Jakob, C.G, Qiu, W, Edalji, R.P, Sun, C. | | Deposit date: | 2016-06-17 | | Release date: | 2017-09-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours.
Nature, 550, 2017
|
|
5KLV
 
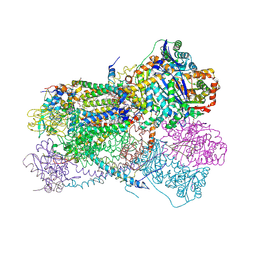 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5KPD
 
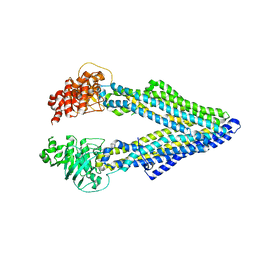 | | Mouse pgp 34 linker deleted double EQ mutant | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-07-03 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
5LD2
 
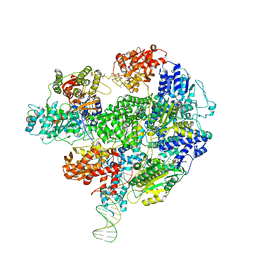 | | Cryo-EM structure of RecBCD+DNA complex revealing activated nuclease domain | | Descriptor: | Fork-Hairpin DNA (70-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Mechanism for nuclease regulation in RecBCD.
Elife, 5, 2016
|
|
5KKN
 
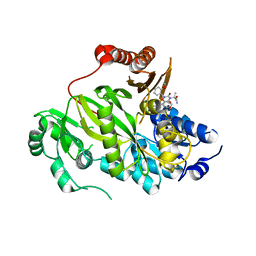 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
5KN9
 
 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5N3W
 
 | | Crystal structure of LTA4H bound to a selective inhibitor against LTB4 generation | | Descriptor: | 3-[2-(2-hydroxyphenyl)ethyl]-5-methoxy-phenol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Wong, C.T, Low, C.M, Snelgrove, R.J, Hare, S.A. | | Deposit date: | 2017-02-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The development of novel LTA4H modulators to selectively target LTB4 generation.
Sci Rep, 7, 2017
|
|
5N6N
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ID3
 
 | | Solution structure of the pore-forming region of C. elegans Mitochondrial Calcium Uniporter (MCU) | | Descriptor: | Mitochondrial Calcium Uniporter | | Authors: | Oxenoid, K, Dong, Y, Cao, C, Cui, T, Sancak, Y, Markhard, A.L, Grabarek, Z, Kong, L, Liu, Z, Ouyang, B, Cong, Y, Mootha, V.K, Chou, J.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Architecture of the mitochondrial calcium uniporter.
Nature, 533, 2016
|
|
