8CR5
 
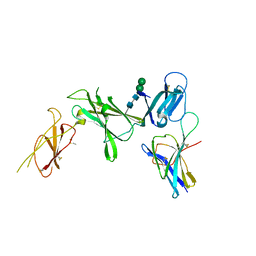 | | Murine Interleukin-12 receptor beta 1 domain 1 in complex with murine Interleukin-12 beta. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5XZH
 
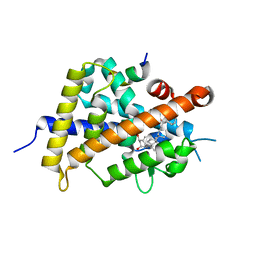 | | Vitamin D receptor with a synthetic ligand ADRO2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6R)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
8E4X
 
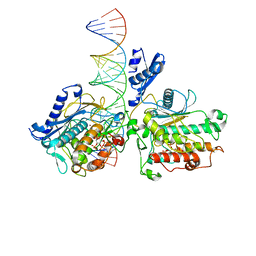 | |
8E0F
 
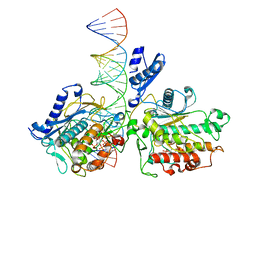 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*GP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site.
Nucleic Acids Res., 50, 2022
|
|
2O4J
 
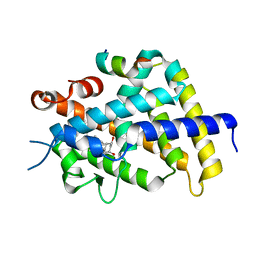 | | Crystal Structure of Rat Vitamin D Receptor Ligand Binding Domain Complexed with VitIII 17-20Z and the NR2 Box of DRIP 205 | | Descriptor: | (1R,3R,7E,17Z)-17-(5-hydroxy-1,5-dimethylhexylidene)-2-methylene-9,10-secoestra-5,7-diene-1,3-diol, Peroxisome proliferator-activated receptor-binding protein, Vitamin D3 receptor | | Authors: | Vanhooke, J.L, Benning, M.M, DeLuca, H.F. | | Deposit date: | 2006-12-04 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | New analogs of 2-methylene-19-nor-(20S)-1,25-dihydroxyvitamin D(3) with conformationally restricted side chains: Evaluation of biological activity and structural determination of VDR-bound conformations.
Arch.Biochem.Biophys., 460, 2007
|
|
2O4R
 
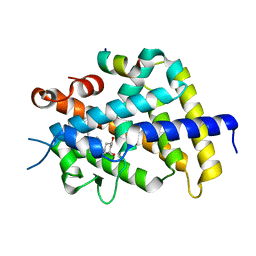 | | Crystal Structure of Rat Vitamin D Receptor Ligand Binding Domain Complexed with VitIII 17-20E and the NR2 Box of DRIP 205 | | Descriptor: | (1R,3R,7E,17E)-17-(5-hydroxy-1,5-dimethylhexylidene)-2-methylene-9,10-secoestra-5,7-diene-1,3-diol, Peroxisome proliferator-activated receptor-binding protein, Vitamin D3 receptor | | Authors: | Vanhooke, J.L, Benning, M.M, DeLuca, H.F. | | Deposit date: | 2006-12-04 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New analogs of 2-methylene-19-nor-(20S)-1,25-dihydroxyvitamin D(3) with conformationally restricted side chains: Evaluation of biological activity and structural determination of VDR-bound conformations.
Arch.Biochem.Biophys., 460, 2007
|
|
7F9K
 
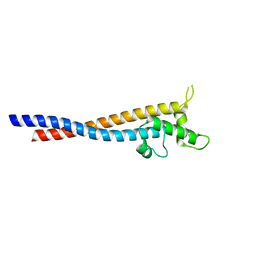 | | Crystal structure of the variable region of Plasmodium RIFIN #6(PF3D7_1400600) | | Descriptor: | Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9N
 
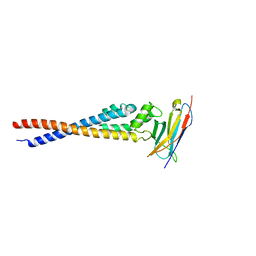 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9L
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #6 (PF3D7_1400600) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9M
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
5VWI
 
 | |
2Q6W
 
 | |
5XS2
 
 | |
5VWC
 
 | | Crystal structure of human Scribble PDZ1 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein scribble homolog | | Authors: | Lim, K.Y.B, Kvansakul, M. | | Deposit date: | 2017-05-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor beta-PIX.
J. Biol. Chem., 292, 2017
|
|
1B3J
 
 | | STRUCTURE OF THE MHC CLASS I HOMOLOG MIC-A, A GAMMADELTA T CELL LIGAND | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I HOMOLOG MIC-A | | Authors: | Li, P, Willie, S, Bauer, S, Morris, D, Spies, T, Strong, R. | | Deposit date: | 1998-12-11 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand.
Immunity, 10, 1999
|
|
2R35
 
 | |
2R34
 
 | |
2R36
 
 | | Crystal structure of ni human ARG-insulin | | Descriptor: | CHLORIDE ION, Insulin, NICKEL (II) ION, ... | | Authors: | Sreekanth, R, Pattabhi, V, Rajan, S.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal induced structural changes observed in hexameric insulin
Int.J.Biol.Macromol., 44, 2009
|
|
6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
7QS8
 
 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | Protein Tax-1, Protein scribble homolog | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2022-01-13 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
7QRS
 
 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | ACETATE ION, GLYCEROL, Protein Tax-1, ... | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
7QRT
 
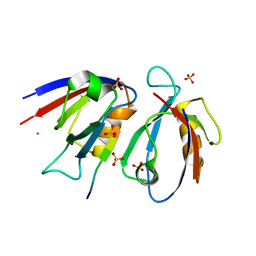 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | CHLORIDE ION, Protein Tax-1, Protein scribble homolog, ... | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M, Maddumage, J.C. | | Deposit date: | 2022-01-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
7QS9
 
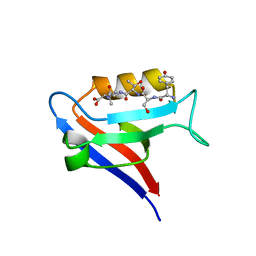 | |
7QSA
 
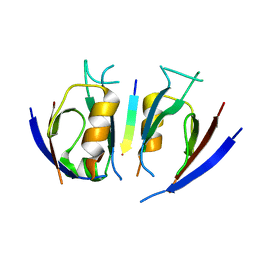 | |
7QSB
 
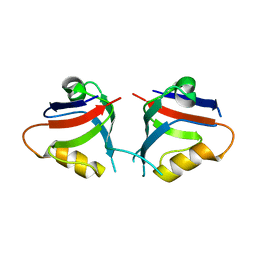 | |
