6A9W
 
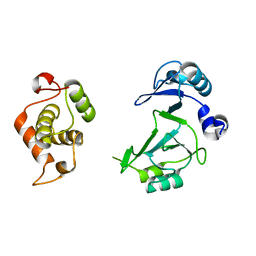 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
2V3L
 
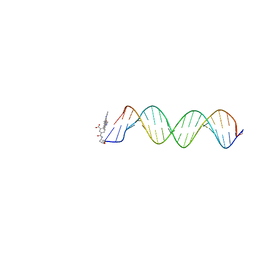 | | Orientational and dynamical heterogeneity of Rhodamine 6G terminally attached to a DNA helix | | Descriptor: | 5'-D(*CP*AP*AP*AP*GP*CP*GP*CP*CP*AP *TP*TP*CP*GP*CP*CP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*GP*GP*CP*GP*AP*AP *TP*GP*GP*CP*GP*CP*TP*TP*TP*G)-3', RHODAMINE 6G | | Authors: | Neubauer, H, Gaiko, N, Berger, S, Schaffer, J, Eggeling, C, Tuma, J, Verdier, L, Seidel, C.A.M, Griesinger, C, Volkmer, A. | | Deposit date: | 2007-06-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Orientational and Dynamical Heterogeneity of Rhodamine 6G Terminally Attached to a DNA Helix Revealed by NMR and Single-Molecule Fluorescence Spectroscopy.
J.Am.Chem.Soc., 129, 2007
|
|
2ODH
 
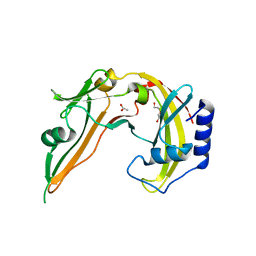 | | Restriction Endonuclease BCNI in the Absence of DNA | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, R.BcnI | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
3WTF
 
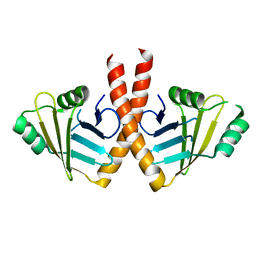 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | Descriptor: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Marsh, M.E, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
3PFI
 
 | | 2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase ruvB | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate
To be Published
|
|
7ZIE
 
 | |
2IVH
 
 | | Crystal structure of the nuclease domain of ColE7 (H545Q mutant) in complex with an 18-bp duplex DNA | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*TP*CP*GP*AP*TP *CP*GP*AP*AP*TP*TP*CP*C)-3', COLCIN-E7, ZINC ION | | Authors: | Wang, Y.T, Yang, W.J, Li, C.L, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2006-06-13 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Sequence-Dependent DNA Cleavage by Nonspecific Endonucleases.
Nucleic Acids Res., 35, 2007
|
|
1VRO
 
 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
6PZ3
 
 | |
6TW3
 
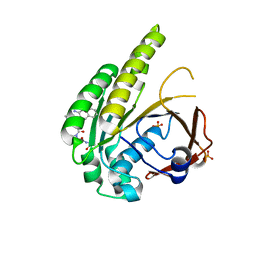 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | Descriptor: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8G1I
 
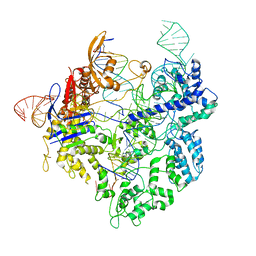 | | SpCas9 with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA E2_NTS, DNA E2_TS, ... | | Authors: | Korolev, S, Gagnon, K. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | SpCas9 with sgRNA and target DNA
To Be Published
|
|
4BLG
 
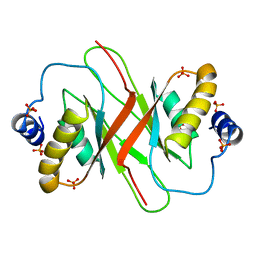 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
8T7S
 
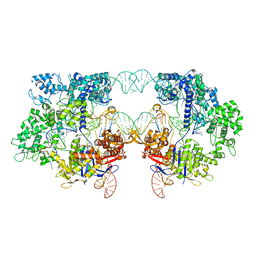 | | SpRYmer bound to NAC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
3QF7
 
 | |
3LHK
 
 | |
3LPV
 
 | | X-ray crystal structure of duplex DNA containing a cisplatin 1,2-d(GpG) intrastrand cross-link | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', Cisplatin, ... | | Authors: | Todd, R.C, Lippard, S.J. | | Deposit date: | 2010-02-06 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of duplex DNA containing the cisplatin 1,2-{Pt(NH(3))(2)}(2+)-d(GpG) cross-link at 1.77A resolution.
J.Inorg.Biochem., 104, 2010
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6V5K
 
 | |
6Q6R
 
 | | Recognition of different base tetrads by RHAU: X-ray crystal structure of G4 recognition motif bound to the 3-end tetrad of a DNA G-quadruplex | | Descriptor: | ATP-dependent DNA/RNA helicase DHX36, POTASSIUM ION, Parallel stranded DNA G-quadruplex | | Authors: | Heddi, B, Cheong, V.V, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex.
J.Struct.Biol., 209, 2020
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
