1OYF
 
 | | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol | | Descriptor: | 6-METHYLHEPTAN-1-OL, ACETIC ACID, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-04-04 | | Release date: | 2003-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol
To be Published
|
|
4U24
 
 | | Crystal structure of the E. coli ribosome bound to dalfopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U85
 
 | | Human Pin1 with cysteine sulfinic acid 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
2H9M
 
 | | WDR5 in complex with unmodified H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9N
 
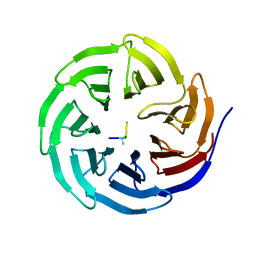 | | WDR5 in complex with monomethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
1P22
 
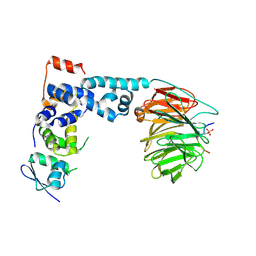 | | Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity on the SCFbeta-TrCP1 ubiquitin ligase | | Descriptor: | Beta-catenin, F-box/WD-repeat protein 1A, Skp1 | | Authors: | Wu, G, Xu, G, Schulman, B.A, Jeffrey, P.D, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a beta-TrCP1-Skp1-beta-Catenin complex: destruction motif binding and lysine specificity of the SCFbeta-TrCP1 ubiquitin ligase
Mol.Cell, 11, 2003
|
|
1TL7
 
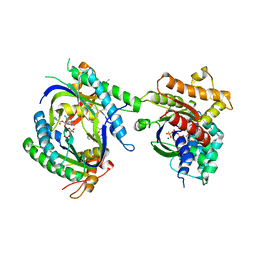 | | Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With 2'(3')-O-(N-methylanthraniloyl)-guanosine 5'-triphosphate and Mn | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | Authors: | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
1SVK
 
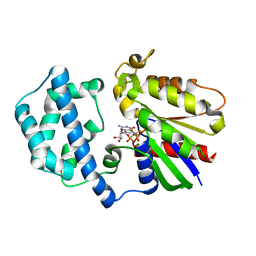 | | Structure of the K180P mutant of Gi alpha subunit bound to AlF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2H9P
 
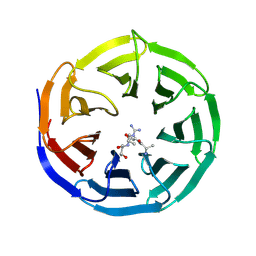 | | WDR5 in complex with trimethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2HES
 
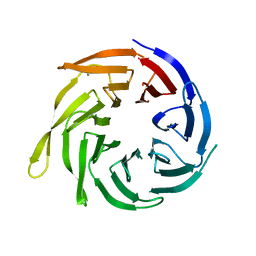 | | Cytosolic Iron-sulphur Assembly Protein- 1 | | Descriptor: | CALCIUM ION, Ydr267cp | | Authors: | Srinivasan, V, Michel, H, Lill, R, Daili, J.A.N, Pierik, A.J. | | Deposit date: | 2006-06-22 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Yeast WD40 Domain Protein Cia1, a Component Acting Late in Iron-Sulfur Protein Biogenesis.
Structure, 15, 2007
|
|
1PFO
 
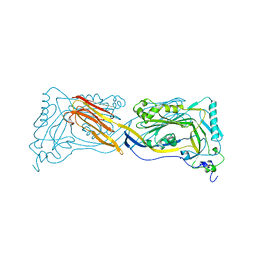 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
1PB5
 
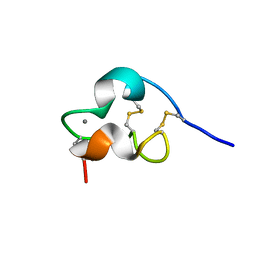 | | NMR Structure of a Prototype LNR Module from Human Notch1 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Vardar, D, North, C.L, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Prototype Lin12-Notch Repeat Module from Human Notch1
Biochemistry, 42, 2003
|
|
1TBG
 
 | | BETA-GAMMA DIMER OF THE HETEROTRIMERIC G-PROTEIN TRANSDUCIN | | Descriptor: | TRANSDUCIN | | Authors: | Sondek, J.S, Bohm, A, Lambright, D.G, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1996-06-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a G-protein beta gamma dimer at 2.1A resolution.
Nature, 379, 1996
|
|
1TQ0
 
 | | Crystal structure of the potent anticoagulant thrombin mutant W215A/E217A in free form | | Descriptor: | Prothrombin | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1TRS
 
 | |
1UB2
 
 | |
1TQ7
 
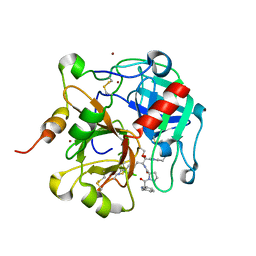 | | Crystal structure of the anticoagulant thrombin mutant W215A/E217A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1TRJ
 
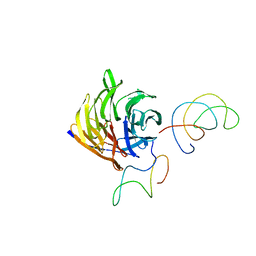 | | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome | | Descriptor: | Guanine nucleotide-binding protein beta subunit-like protein, Helix 39 of 18S rRNA, Helix 40 of 18S rRNA | | Authors: | Sengupta, J, Nilsson, J, Gursky, R, Spahn, C.M, Nissen, P, Frank, J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Nat.Struct.Mol.Biol., 11, 2004
|
|
2HL7
 
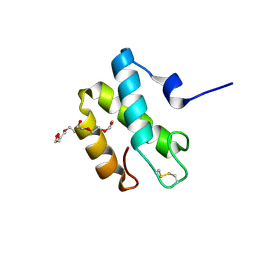 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | Descriptor: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | Authors: | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | Deposit date: | 2006-07-06 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
2I1X
 
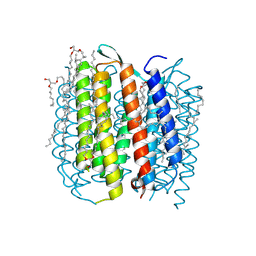 | | Bacteriorhodopsin/lipid complex, D96A mutant | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, ... | | Authors: | Lanyi, J.K, Schobert, B. | | Deposit date: | 2006-08-15 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Propagating Structural Perturbation Inside Bacteriorhodopsin: Crystal Structures of the M State and the D96A and T46V Mutants.
Biochemistry, 45, 2006
|
|
2I94
 
 | | NMR Structure of recoverin bound to rhodopsin kinase | | Descriptor: | CALCIUM ION, Recoverin, Rhodopsin kinase | | Authors: | Ames, J.B. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Calcium-induced Inhibition of Rhodopsin Kinase by Recoverin.
J.Biol.Chem., 281, 2006
|
|
9BQJ
 
 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
3DF0
 
 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
3DAW
 
 | | Structure of the actin-depolymerizing factor homology domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Paavilainen, V.O, Oksanen, E, Goldman, A, Lappalainen, P. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the actin-depolymerizing factor homology domain in complex with actin
J.Cell Biol., 182, 2008
|
|
6U4A
 
 | | BRD3-BD1 in complex with the cyclic peptide 3.1_3 | | Descriptor: | Bromodomain-containing protein 3, GLYCEROL, SULFATE ION, ... | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
